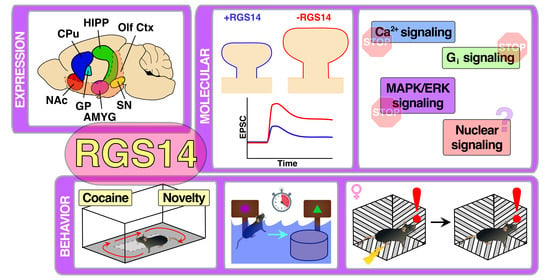
RGS14 Regulation of Post-Synaptic Signaling and Spine Plasticity in Brain
Abstract
:1. Introduction
1.1. RGS Protein Regulation of G-Protein Signaling
1.2. RGS14 Interacting Partners
2. RGS14 Protein Tissue Distribution
3. RGS14 Expression in the Brain
3.1. RGS14 Developmental Expression
3.2. RGS14 Expression in Adult Rodent and Primate Brains
4. RGS14 in the Hippocampus
5. RGS14 Regulation of Postsynaptic Signaling
5.1. RGS14 Regulation of GPCR–Gi/o Activation
5.2. RGS14 Regulation of Inactive Gi1/3
5.3. Regulation of RGS14 Signaling by the 14-3-3 Scaffolding Protein
5.4. RGS14 Regulation of Monomeric GTPase Signaling
5.5. Proteins Involved in Dendritic Spine Structure
5.6. RGS14 Binding to Calmodulin/Calmodulin-Dependent Kinase and Regulation of Spine Calcium
6. RGS14 Regulation of Long-Term Potentiation (LTP)
6.1. RGS14 Regulation of E-LTP and Potential Mechanisms
6.2. Potential Roles for RGS14 in L-LTP and in the Nucleus
6.3. Modulation of RGS14 Inhibition on CA2 Plasticity
7. Roles for RGS14 in Hippocampal Behavior
8. Roles for RGS14 in Non-Hippocampal Brain Regions
8.1. Amygdala
8.2. Ventral Striatum
8.3. Basal Ganglia
8.4. Link to Synaptic Plasticity outside of Hippocampus
8.5. Implications of RGS14 outside of the Hippocampus
9. Impact of Genetic Variation on RGS14 Function and Physiology in Brain
10. Unanswered Questions and Future Directions for the Study of RGS14
10.1. A Nuclear Function for RGS14?
10.2. Modulation of RGS14 Activity
10.3. RGS14 as a PDZ Scaffold Protein in the Brain
10.4. Beyond Learning and Memory: RGS14 outside the Hippocampus
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stewart, A.; Fisher, R.A. Introduction: G Protein-coupled Receptors and RGS Proteins. Prog. Mol. Biol. Transl. Sci. 2015, 133, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kimple, A.J.; Bosch, D.E.; Giguere, P.M.; Siderovski, D.P. Regulators of G-protein signaling and their Gα substrates: Promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 2011, 63, 728–749. [Google Scholar] [CrossRef] [Green Version]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Hollinger, S.; Hepler, J.R. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol. Rev. 2002, 54, 527–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjögren, B. The evolution of regulators of G protein signaling proteins as drug targets—20 years in the making: IUPHAR Review 21. Br. J. Pharmacol. 2017, 174, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Snow, B.E.; Antonio, L.; Suggs, S.; Gutstein, H.B.; Siderovski, D.P. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem. Biophys. Res. Commun. 1997, 233, 770–777. [Google Scholar] [CrossRef]
- Vellano, C.P.; Lee, S.E.; Dudek, S.M.; Hepler, J.R. RGS14 at the interface of hippocampal signaling and synaptic plasticity. Trends Pharmacol. Sci. 2011, 32, 666–674. [Google Scholar] [CrossRef] [Green Version]
- Traver, S.; Bidot, C.; Spassky, N.; Baltauss, T.; De Tand, M.F.; Thomas, J.L.; Zalc, B.; Janoueix-Lerosey, I.; Gunzburg, J.D. RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of Gαo. Biochem. J. 2000, 350 Pt 1, 19–29. [Google Scholar] [CrossRef]
- Hollinger, S.; Taylor, S.B.; Goldman, E.H.; Hepler, J.R. RGS14 is a bifunctional regulator of Gαi/o activity that exists in multiple populations in brain. J. Neurochem. 2001, 79, 941–949. [Google Scholar] [CrossRef]
- Traver, S.; Splingard, A.; Gaudriault, G.; De Gunzburg, J. The RGS (regulator of G-protein signalling) and GoLoco domains of RGS14 co-operate to regulate Gi-mediated signalling. Biochem. J. 2004, 379, 627–632. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.E.; Goswami, D.; Branch, M.R.; Ramineni, S.; Ortlund, E.A.; Griffin, P.R.; Hepler, J.R. Integration of G protein α (Gα) signaling by the regulator of G protein signaling 14 (RGS14). J. Biol. Chem. 2015, 290, 9037–9049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellano, C.P.; Maher, E.M.; Hepler, J.R.; Blumer, J.B. G protein-coupled receptors and resistance to inhibitors of cholinesterase-8A (Ric-8A) both regulate the regulator of g protein signaling 14 RGS14.Gαi1 complex in live cells. J. Biol. Chem. 2011, 286, 38659–38669. [Google Scholar] [CrossRef] [Green Version]
- Vellano, C.P.; Brown, N.E.; Blumer, J.B.; Hepler, J.R. Assembly and function of the regulator of G protein signaling 14 (RGS14).H-Ras signaling complex in live cells are regulated by Gαi1 and Gαi-linked G protein-coupled receptors. J. Biol. Chem. 2013, 288, 3620–3631. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Nunn, C.; Ramineni, S.; Hepler, J.R.; Chidiac, P. The Ras-binding domain region of RGS14 regulates its functional interactions with heterotrimeric G proteins. J. Cell. Biochem. 2013, 114, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Shu, F.J.; Ramineni, S.; Hepler, J.R. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell. Signal. 2010, 22, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Evans, P.R.; Gerber, K.J.; Dammer, E.B.; Duong, D.M.; Goswami, D.; Lustberg, D.J.; Zou, J.; Yang, J.J.; Dudek, S.M.; Griffin, P.R.; et al. Interactome Analysis Reveals Regulator of G Protein Signaling 14 (RGS14) is a Novel Calcium/Calmodulin (Ca2+/CaM) and CaM Kinase II (CaMKII) Binding Partner. J. Proteome Res. 2018, 17, 1700–1711. [Google Scholar] [CrossRef]
- Gerber, K.J.; Squires, K.E.; Hepler, J.R. 14-3-3γ binds regulator of G protein signaling 14 (RGS14) at distinct sites to inhibit the RGS14:Gαi-AlF4− signaling complex and RGS14 nuclear localization. J. Biol. Chem. 2018, 293, 14616–14631. [Google Scholar] [CrossRef] [Green Version]
- Squires, K.E.; Gerber, K.J.; Tillman, M.C.; Lustberg, D.J.; Montanez-Miranda, C.; Zhao, M.; Ramineni, S.; Scharer, C.D.; Saha, R.N.; Shu, F.J.; et al. Human genetic variants disrupt RGS14 nuclear shuttling and regulation of LTP in hippocampal neurons. J. Biol. Chem. 2020, 296, 100024. [Google Scholar] [CrossRef]
- Shu, F.J.; Ramineni, S.; Amyot, W.; Hepler, J.R. Selective interactions between Giα1 and Giα3 and the GoLoco/GPR domain of RGS14 influence its dynamic subcellular localization. Cell. Signal. 2007, 19, 163–176. [Google Scholar] [CrossRef]
- Cho, H.; Kim, D.U.; Kehrl, J.H. RGS14 is a centrosomal and nuclear cytoplasmic shuttling protein that traffics to promyelocytic leukemia nuclear bodies following heat shock. J. Biol. Chem. 2005, 280, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Vatner, D.E.; Zhang, J.; Oydanich, M.; Guers, J.; Katsyuba, E.; Yan, L.; Sinclair, D.; Auwerx, J.; Vatner, S.F. Enhanced longevity and metabolism by brown adipose tissue with disruption of the regulator of G protein signaling 14. Aging Cell 2018, 17, e12751. [Google Scholar] [CrossRef]
- Homologene. Regulator of G-Protein Signaling 14. Available online: https://www.ncbi.nlm.nih.gov/homologene/4735 (accessed on 18 April 2021).
- Kardestuncer, T.; Wu, H.; Lim, A.L.; Neer, E.J. Cardiac myocytes express mRNA for ten RGS proteins: Changes in RGS mRNA expression in ventricular myocytes and cultured atria. FEBS Lett. 1998, 438, 285–288. [Google Scholar] [CrossRef] [Green Version]
- Agudelo, L.Z.; Ferreira, D.M.S.; Cervenka, I.; Bryzgalova, G.; Dadvar, S.; Jannig, P.R.; Pettersson-Klein, A.T.; Lakshmikanth, T.; Sustarsic, E.G.; Porsmyr-Palmertz, M.; et al. Kynurenic Acid and Gpr35 Regulate Adipose Tissue Energy Homeostasis and Inflammation. Cell Metab. 2018, 27, 378–392.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squires, K.E.; Gerber, K.J.; Pare, J.F.; Branch, M.R.; Smith, Y.; Hepler, J.R. Regulator of G protein signaling 14 (RGS14) is expressed pre- and postsynaptically in neurons of hippocampus, basal ganglia, and amygdala of monkey and human brain. Brain Struct. Funct. 2018, 223, 233–253. [Google Scholar] [CrossRef]
- Evans, P.R.; Lee, S.E.; Smith, Y.; Hepler, J.R. Postnatal developmental expression of regulator of G protein signaling 14 (RGS14) in the mouse brain. J. Comp. Neurol. 2014, 522, 186–203. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Li, Y.; Tang, X.H.; Li, X.H.; Dai, H.J.; Miao, R.J.; Cai, J.J.; Huang, Z.J.; Chen, A.F.; Xing, X.W.; Lu, Y.; et al. Regulator of G protein signalling 14 attenuates cardiac remodelling through the MEK-ERK1/2 signalling pathway. Basic Res. Cardiol. 2016, 111, 47. [Google Scholar] [CrossRef] [Green Version]
- Mittmann, C.; Chung, C.H.; Hoppner, G.; Michalek, C.; Nose, M.; Schuler, C.; Schuh, A.; Eschenhagen, T.; Weil, J.; Pieske, B.; et al. Expression of ten RGS proteins in human myocardium: Functional characterization of an upregulation of RGS4 in heart failure. Cardiovasc. Res. 2002, 55, 778–786. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Mende, U. Regulators of G-protein signaling in the heart and their potential as therapeutic targets. Circ. Res. 2011, 109, 320–333. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Rodan, A.R.; Le, T.H.; Gaulton, K.J.; Haessler, J.; Stilp, A.M.; Kamatani, Y.; Zhu, G.; Sofer, T.; Puri, S.; et al. Trans-ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity. Am. J. Hum. Genet. 2016, 99, 636–646. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.C.; Chou, W.H.; Chu, H.W.; Huang, C.C.; Liu, X.; Chang, W.P.; Chou, Y.H.; Chang, W.C. The rs1256328 (ALPL) and rs12654812 (RGS14) Polymorphisms are Associated with Susceptibility to Calcium Nephrolithiasis in a Taiwanese population. Sci. Rep. 2019, 9, 17296. [Google Scholar] [CrossRef] [Green Version]
- Urabe, Y.; Tanikawa, C.; Takahashi, A.; Okada, Y.; Morizono, T.; Tsunoda, T.; Kamatani, N.; Kohri, K.; Chayama, K.; Kubo, M.; et al. A genome-wide association study of nephrolithiasis in the Japanese population identifies novel susceptible Loci at 5q35.3, 7p14.3, and 13q14.1. PLoS Genet. 2012, 8, e1002541. [Google Scholar] [CrossRef] [Green Version]
- Yasui, T.; Okada, A.; Urabe, Y.; Usami, M.; Mizuno, K.; Kubota, Y.; Tozawa, K.; Sasaki, S.; Higashi, Y.; Sato, Y.; et al. A replication study for three nephrolithiasis loci at 5q35.3, 7p14.3 and 13q14.1 in the Japanese population. J. Hum. Genet. 2013, 58, 588–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.; Chen, Y.; Lin, H.; Liao, M.; Li, T.; Tong, L.; Wei, S.; Xian, X.; Zhu, J.; Chen, J.; et al. Significant association between RGS14 rs12654812 and nephrolithiasis risk among Guangxi population in China. J. Clin. Lab. Anal. 2018, 32, e22435. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Han, W.; Ni, T.; Zhao, L.; Li, X.; Zhang, B.; Zhang, T. Genetic Polymorphisms of RGS14 and Renal Stone Disease: A Case-control Study Based on the Chinese Han Population. Arch. Med. Res. 2020, 52, 332–338. [Google Scholar] [CrossRef]
- Kestenbaum, B.; Glazer, N.L.; Kottgen, A.; Felix, J.F.; Hwang, S.J.; Liu, Y.; Lohman, K.; Kritchevsky, S.B.; Hausman, D.B.; Petersen, A.K.; et al. Common genetic variants associate with serum phosphorus concentration. J. Am. Soc. Nephrol. 2010, 21, 1223–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, R.B.; Razzaque, M.S. Dysregulation of phosphate metabolism and conditions associated with phosphate toxicity. BoneKEy Rep. 2015, 4, 705. [Google Scholar] [CrossRef] [Green Version]
- Robinson-Cohen, C.; Lutsey, P.L.; Kleber, M.E.; Nielson, C.M.; Mitchell, B.D.; Bis, J.C.; Eny, K.M.; Portas, L.; Eriksson, J.; Lorentzon, M.; et al. Genetic Variants Associated with Circulating Parathyroid Hormone. J. Am. Soc. Nephrol. 2017, 28, 1553–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson-Cohen, C.; Bartz, T.M.; Lai, D.; Ikizler, T.A.; Peacock, M.; Imel, E.A.; Michos, E.D.; Foroud, T.M.; Akesson, K.; Taylor, K.D.; et al. Genetic Variants Associated with Circulating Fibroblast Growth Factor 23. J. Am. Soc. Nephrol. 2018, 29, 2583–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silver, J.; Rodriguez, M.; Slatopolsky, E. FGF23 and PTH—Double agents at the heart of CKD. Nephrol. Dial. Transplant. 2012, 27, 1715–1720. [Google Scholar] [CrossRef] [PubMed]
- Friedman, P.A.; Mamonova, T.; Magyar, C.E.; Squires, K.E.; Sneddon, W.B.; Emlet, D.R.; Hepler, J.R. Genetic variants disrupt human RGS14 binding to NHERF1 and regulation of NPT2A-mediated phosphate transport. bioRxiv 2019. [Google Scholar] [CrossRef]
- Reif, K.; Cyster, J.G. RGS molecule expression in murine B lymphocytes and ability to down-regulate chemotaxis to lymphoid chemokines. J. Immunol. 2000, 164, 4720–4729. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Kozasa, T.; Takekoshi, K.; De Gunzburg, J.; Kehrl, J.H. RGS14, a GTPase-Activating Protein for Gia, Attenuates Gia- and G13a-mediated signaling pathways. Mol. Pharmacol. 2000, 58, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Beadling, C.; Druey, K.M.; Richter, G.; Kehrl, J.H.; Smith, K.A. Regulators of G protein signaling exhibit distinct patterns of gene expression and target G protein specificity in human lymphocytes. J. Immunol. 1999, 162, 2677–2682. [Google Scholar]
- Lim, J.; Thompson, J.; May, R.C.; Hotchin, N.A.; Caron, E. Regulator of G-Protein Signalling-14 (RGS14) Regulates the Activation of αMβ2 Integrin during Phagocytosis. PLoS ONE 2013, 8, e69163. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 46, D1271–D1281. [Google Scholar] [CrossRef]
- Smith, C.M.; Hayamizu, T.F.; Finger, J.H.; Bello, S.M.; McCright, I.J.; Xu, J.; Baldarelli, R.M.; Beal, J.S.; Campbell, J.; Corbani, L.E.; et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019, 47, D774–D779. [Google Scholar] [CrossRef] [Green Version]
- Craig, R.; Cortens, J.P.; Beavis, R.C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004, 3, 1234–1242. [Google Scholar] [CrossRef]
- Hawrylycz, M.J.; Lein, E.S.; Guillozet-Bongaarts, A.L.; Shen, E.H.; Ng, L.; Miller, J.A.; van de Lagemaat, L.N.; Smith, K.A.; Ebbert, A.; Riley, Z.L.; et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012, 489, 391–399. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Foster, S.L.; Lustberg, D.J.; Harbin, N.H.; Bramlett, S.N.; Hepler, J.R.; Weinshenker, D. RGS14 modulates locomotor behavior and ERK signaling induced by environmental novelty and cocaine within discrete limbic structures. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lee, S.E.; Simons, S.B.; Heldt, S.A.; Zhao, M.L.; Schroeder, J.P.; Vellano, C.P.; Cowan, D.P.; Ramineni, S.; Yates, C.K.; Feng, Y.; et al. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. USA 2010, 107, 16994–16998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, C.M.; Burgess, N. The hippocampus and memory: Insights from spatial processing. Nat. Rev. Neurosci. 2008, 9, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Tzakis, N.; Holahan, M.R. Social Memory and the Role of the Hippocampal CA2 Region. Front. Behav. Neurosci. 2019, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.; Amaral, D.; Morris, R.; Bliss, T.; O’Keefe, J. The Hippocampus Book; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Basu, J.; Siegelbaum, S.A. The Corticohippocampal Circuit, Synaptic Plasticity, and Memory. Cold Spring Harb. Perspect. Biol. 2015, 7, a021733. [Google Scholar] [CrossRef] [PubMed]
- Middleton, S.J.; McHugh, T.J. CA2: A Highly Connected Intrahippocampal Relay. Annu. Rev. Neurosci. 2020, 43, 55–72. [Google Scholar] [CrossRef]
- Robert, V.; Cassim, S.; Chevaleyre, V.; Piskorowski, R.A. Hippocampal area CA2: Properties and contribution to hippocampal function. Cell Tissue Res. 2018, 373, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Chevaleyre, V.; Piskorowski, R.A. Hippocampal Area CA2: An Overlooked but Promising Therapeutic Target. Trends Mol. Med. 2016, 22, 645–655. [Google Scholar] [CrossRef]
- Carstens, K.E.; Dudek, S.M. Regulation of synaptic plasticity in hippocampal area CA2. Curr. Opin. Neurobiol. 2019, 54, 194–199. [Google Scholar] [CrossRef]
- Dudek, S.M.; Alexander, G.M.; Farris, S. Rediscovering area CA2: Unique properties and functions. Nat. Rev. Neurosci. 2016, 17, 89–102. [Google Scholar] [CrossRef] [Green Version]
- Chevaleyre, V.; Siegelbaum, S.A. Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico-hippocampal loop. Neuron 2010, 66, 560–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hitti, F.L.; Siegelbaum, S.A. The hippocampal CA2 region is essential for social memory. Nature 2014, 508, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Laham, B.J.; Diethorn, E.J.; Gould, E. Newborn mice form lasting CA2-dependent memories of their mothers. Cell Rep. 2021, 34, 108668. [Google Scholar] [CrossRef]
- Mankin, E.A.; Diehl, G.W.; Sparks, F.T.; Leutgeb, S.; Leutgeb, J.K. Hippocampal CA2 activity patterns change over time to a larger extent than between spatial contexts. Neuron 2015, 85, 190–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacDonald, C.J.; Tonegawa, S. Crucial role for CA2 inputs in the sequential organization of CA1 time cells supporting memory. Proc. Natl. Acad. Sci. USA 2021, 118, e2020698118. [Google Scholar] [CrossRef] [PubMed]
- Häussler, U.; Rinas, K.; Kilias, A.; Egert, U.; Haas, C.A. Mossy fiber sprouting and pyramidal cell dispersion in the hippocampal CA2 region in a mouse model of temporal lobe epilepsy. Hippocampus 2016, 26, 577–588. [Google Scholar] [CrossRef]
- Boehringer, R.; Polygalov, D.; Huang, A.J.Y.; Middleton, S.J.; Robert, V.; Wintzer, M.E.; Piskorowski, R.A.; Chevaleyre, V.; McHugh, T.J. Chronic Loss of CA2 Transmission Leads to Hippocampal Hyperexcitability. Neuron 2017, 94, 642–655.e9. [Google Scholar] [CrossRef] [Green Version]
- Leroy, F.; Brann, D.H.; Meira, T.; Siegelbaum, S.A. Input-Timing-Dependent Plasticity in the Hippocampal CA2 Region and Its Potential Role in Social Memory. Neuron 2017, 95, 1089–1102.e5. [Google Scholar] [CrossRef]
- Dominguez, S.; Rey, C.C.; Therreau, L.; Fanton, A.; Massotte, D.; Verret, L.; Piskorowski, R.A.; Chevaleyre, V. Maturation of PNN and ErbB4 Signaling in Area CA2 during Adolescence Underlies the Emergence of PV Interneuron Plasticity and Social Memory. Cell Rep. 2019, 29, 1099–1112.e4. [Google Scholar] [CrossRef] [Green Version]
- Modi, B.; Pimpinella, D.; Pazienti, A.; Zacchi, P.; Cherubini, E.; Griguoli, M. Possible Implication of the CA2 Hippocampal Circuit in Social Cognition Deficits Observed in the Neuroligin 3 Knock-Out Mouse, a Non-Syndromic Animal Model of Autism. Front. Psychiatry 2019, 10, 513. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Choi, Y.S.; Obrietan, K.; Dudek, S.M. Synaptic plasticity (and the lack thereof) in hippocampal CA2 neurons. J. Neurosci. 2007, 27, 12025–12032. [Google Scholar] [CrossRef]
- Simons, S.B.; Escobedo, Y.; Yasuda, R.; Dudek, S.M. Regional differences in hippocampal calcium handling provide a cellular mechanism for limiting plasticity. Proc. Natl. Acad. Sci. USA 2009, 106, 14080–14084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P.R.; Parra-Bueno, P.; Smirnov, M.S.; Lustberg, D.J.; Dudek, S.M.; Hepler, J.R.; Yasuda, R. RGS14 Restricts Plasticity in Hippocampal CA2 by Limiting Postsynaptic Calcium Signaling. eNeuro 2018, 5, ENEURO.0353-17.2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piskorowski, R.A.; Chevaleyre, V. Delta-opioid receptors mediate unique plasticity onto parvalbumin-expressing interneurons in area CA2 of the hippocampus. J. Neurosci. 2013, 33, 14567–14578. [Google Scholar] [CrossRef]
- Nasrallah, K.; Therreau, L.; Robert, V.; Huang, A.J.Y.; McHugh, T.J.; Piskorowski, R.A.; Chevaleyre, V. Routing Hippocampal Information Flow through Parvalbumin Interneuron Plasticity in Area CA2. Cell Rep. 2019, 27, 86–98.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carstens, K.E.; Phillips, M.L.; Pozzo-Miller, L.; Weinberg, R.J.; Dudek, S.M. Perineuronal Nets Suppress Plasticity of Excitatory Synapses on CA2 Pyramidal Neurons. J. Neurosci. 2016, 36, 6312–6320. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, A.; Matsumoto, N.; Morikawa, S.; Tamura, H.; Ikegaya, Y. Juvenile Hippocampal CA2 Region Expresses Aggrecan. Front. Neuroanat. 2017, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelkey, K.A.; Askalan, R.; Paul, S.; Kalia, L.V.; Nguyen, T.H.; Pitcher, G.M.; Salter, M.W.; Lombroso, P.J. Tyrosine phosphatase STEP is a tonic brake on induction of long-term potentiation. Neuron 2002, 34, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Hensch, T.K. Critical period regulation. Annu. Rev. Neurosci. 2004, 27, 549–579. [Google Scholar] [CrossRef] [Green Version]
- Patton, M.H.; Blundon, J.A.; Zakharenko, S.S. Rejuvenation of plasticity in the brain: Opening the critical period. Curr. Opin. Neurobiol. 2019, 54, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Honkura, N.; Ellis-Davies, G.C.; Kasai, H. Structural basis of long-term potentiation in single dendritic spines. Nature 2004, 429, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Shi, S.H.; Esteban, J.A.; Piccini, A.; Poncer, J.C.; Malinow, R. Driving AMPA Receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science 2000, 287, 2262–2267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsien, J.Z.; Huerta, P.T.; Tonegawa, S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell 1996, 87, 1327–1338. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Carew, T.J. Small G protein signaling in neuronal plasticity and memory formation: The specific role of ras family proteins. Neuron 2010, 68, 340–361. [Google Scholar] [CrossRef] [Green Version]
- Herring, B.E.; Nicoll, R.A. Long-Term Potentiation: From CaMKII to AMPA Receptor Trafficking. Annu. Rev. Physiol. 2016, 78, 351–365. [Google Scholar] [CrossRef]
- Gerber, K.J.; Squires, K.E.; Hepler, J.R. Roles for Regulator of G Protein Signaling Proteins in Synaptic Signaling and Plasticity. Mol. Pharmacol. 2016, 89, 273–286. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, Y. 14-3-3 Proteins in Glutamatergic Synapses. Neural Plast. 2018, 2018, 8407609. [Google Scholar] [CrossRef]
- De Oliveira, P.G.; Ramos, M.L.S.; Amaro, A.J.; Dias, R.A.; Vieira, S.I. Gi/o-Protein Coupled Receptors in the Aging Brain. Front. Aging Neurosci. 2019, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, A.; Lim, Y.J.; Kumar, K.; Baby, N.; Pang, K.L.K.; Benoy, A.; Behnisch, T.; Sajikumar, S. Group III metabotropic glutamate receptors gate long-term potentiation and synaptic tagging/capture in rat hippocampal area CA2. eLife 2020, 9, e55344. [Google Scholar] [CrossRef] [PubMed]
- Manvich, D.F.; Petko, A.K.; Branco, R.C.; Foster, S.L.; Porter-Stransky, K.A.; Stout, K.A.; Newman, A.H.; Miller, G.W.; Paladini, C.A.; Weinshenker, D. Selective D2 and D3 receptor antagonists oppositely modulate cocaine responses in mice via distinct postsynaptic mechanisms in nucleus accumbens. Neuropsychopharmacology 2019, 44, 1445–1455. [Google Scholar] [CrossRef]
- Zaniewska, M.; Filip, M.; Przegalinski, E. The Involvement of Norepinephrine in Behaviors Related to Psychostimulant Addiction. Curr. Neuropharmacol. 2015, 13, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Branch, M.R.; Hepler, J.R. Endogenous RGS14 is a cytoplasmic-nuclear shuttling protein that localizes to juxtanuclear membranes and chromatin-rich regions of the nucleus. PLoS ONE 2017, 12, e0184497. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.E.; Lambert, N.A.; Hepler, J.R. RGS14 regulates the lifetime of Gα-GTP signaling but does not prolong Gβγsignaling following receptor activation in live cells. Pharmacol. Res. Perspect. 2016, 4, e00249. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.R.; Parikh, H.; Park, Y. Loco signaling pathway in longevity. Small GTPases 2011, 2, 158–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, H.; Foote, M.; Graham, K.; Wu, Y.; Zhou, Y. 14-3-3 proteins are required for hippocampal long-term potentiation and associative learning and memory. J. Neurosci. 2014, 34, 4801–4808. [Google Scholar] [CrossRef]
- Willard, F.S.; Willard, M.D.; Kimple, A.J.; Soundararajan, M.; Oestreich, E.A.; Li, X.; Sowa, N.A.; Kimple, R.J.; Doyle, D.A.; Der, C.J.; et al. Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS ONE 2009, 4, e4884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Pak, D.; Qin, Y.; McCormack, S.G.; Kim, M.J.; Baumgart, J.P.; Velamoor, V.; Auberson, Y.P.; Osten, P.; van Aelst, L.; et al. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron 2005, 46, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Mittal, V.; Linder, M.E. Biochemical characterization of RGS14: RGS14 activity towards G-protein α subunits is independent of its binding to Rap2A. Biochem. J. 2006, 394, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Kneussel, M.; Wagner, W. Myosin motors at neuronal synapses: Drivers of membrane transport and actin dynamics. Nat. Rev. Neurosci. 2013, 14, 233–247. [Google Scholar] [CrossRef]
- Koleske, A.J. Molecular mechanisms of dendrite stability. Nat. Rev. Neurosci. 2013, 14, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Dunn, H.A.; Ferguson, S.S. PDZ Protein Regulation of G Protein-Coupled Receptor Trafficking and Signaling Pathways. Mol. Pharmacol. 2015, 88, 624–639. [Google Scholar] [CrossRef] [Green Version]
- Woolfrey, K.M.; Dell’Acqua, M.L. Coordination of Protein Phosphorylation and Dephosphorylation in Synaptic Plasticity. J. Biol. Chem. 2015, 290, 28604–28612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.; Liu, L.; Wong, T.P.; Wu, D.C.; Burette, A.; Weinberg, R.; Wang, Y.T.; Sheng, M. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron 2006, 49, 175–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rex, C.S.; Gavin, C.F.; Rubio, M.D.; Kramar, E.A.; Chen, L.Y.; Jia, Y.; Huganir, R.L.; Muzyczka, N.; Gall, C.M.; Miller, C.A.; et al. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron 2010, 67, 603–617. [Google Scholar] [CrossRef] [Green Version]
- Wagner, W.; Brenowitz, S.D.; Hammer, J.A., 3rd. Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat. Cell Biol. 2011, 13, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Osterweil, E.; Wells, D.G.; Mooseker, M.S. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J. Cell Biol. 2005, 168, 329–338. [Google Scholar] [CrossRef]
- Nakahata, Y.; Yasuda, R. Plasticity of Spine Structure: Local Signaling, Translation and Cytoskeletal Reorganization. Front. Synaptic Neurosci. 2018, 10, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-McCaffrey, L.; Willard, F.S.; Pajak, A.; Dagnino, L.; Siderovski, D.P.; D’Souza, S.J. RGS14 is a microtubule-associated protein. Cell Cycle 2005, 4, 953–960. [Google Scholar] [CrossRef] [Green Version]
- Sheng, M.; Kim, E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 2011, 3, a005678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.K.; Tang, C.; Verpelli, C.; Narayanan, R.; Stearns, M.H.; Xu, R.M.; Li, H.; Sala, C.; Hayashi, Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 2009, 137, 159–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Montañez, E.; Acevedo, M.J.; Lopez-Tellez, J.F.; Duncan, R.S.; Mateos, A.G.; Pavia, J.; Koulen, P.; Khan, Z.U. Regulator of G-protein signaling 14 protein modulates Ca2+ influx through Cav1 channels. NeuroReport 2010, 21, 1034–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Ehlers, M.D.; Bernhardt, J.P.; Su, C.T.; Huganir, R.L. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron 1998, 21, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Ben-Johny, M.; Yue, D.T. Calmodulin regulation (calmodulation) of voltage-gated calcium channels. J. Gen. Physiol. 2014, 143, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Sanhueza, M.; Fernandez-Villalobos, G.; Stein, I.S.; Kasumova, G.; Zhang, P.; Bayer, K.U.; Otmakhov, N.; Hell, J.W.; Lisman, J. Role of the CaMKII/NMDA receptor complex in the maintenance of synaptic strength. J. Neurosci. 2011, 31, 9170–9178. [Google Scholar] [CrossRef] [Green Version]
- Kostic, S.; Pan, B.; Guo, Y.; Yu, H.; Sapunar, D.; Kwok, W.M.; Hudmon, A.; Wu, H.E.; Hogan, Q.H. Regulation of voltage-gated Ca2+ currents by Ca2+/calmodulin-dependent protein kinase II in resting sensory neurons. Mol. Cell. Neurosci. 2014, 62, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Wayman, G.A.; Lee, Y.S.; Tokumitsu, H.; Silva, A.J.; Soderling, T.R. Calmodulin-kinases: Modulators of neuronal development and plasticity. Neuron 2008, 59, 914–931. [Google Scholar] [CrossRef] [Green Version]
- Colbran, R.J.; Brown, A.M. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr. Opin. Neurobiol. 2004, 14, 318–327. [Google Scholar] [CrossRef]
- Okamoto, K.; Bosch, M.; Hayashi, Y. The roles of CaMKII and F-actin in the structural plasticity of dendritic spines: A potential molecular identity of a synaptic tag? Physiology 2009, 24, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Lisman, J.; Yasuda, R.; Raghavachari, S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012, 13, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Baltaci, S.B.; Mogulkoc, R.; Baltaci, A.K. Molecular Mechanisms of Early and Late LTP. Neurochem. Res. 2019, 44, 281–296. [Google Scholar] [CrossRef]
- Bliss, T.V.; Collingridge, G.L. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature 1993, 361, 31–39. [Google Scholar] [CrossRef]
- Lee, S.J.; Escobedo-Lozoya, Y.; Szatmari, E.M.; Yasuda, R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 2009, 458, 299–304. [Google Scholar] [CrossRef]
- Barria, A.; Malinow, R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron 2005, 48, 289–301. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.H.; Hayashi, Y.; Petralia, R.S.; Zaman, S.H.; Wenthold, R.J.; Svoboda, K.; Malinow, R. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 1999, 284, 1811–1816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, H.; Malinow, R. AMPA receptor incorporation into synapses during LTP: The role of lateral movement and exocytosis. Neuron 2009, 64, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, A.C.; Nicoll, R.A. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 2011, 70, 178–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penzes, P.; Cahill, M.E.; Jones, K.A.; Srivastava, D.P. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008, 18, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, L.A.; Goda, Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008, 9, 344–356. [Google Scholar] [CrossRef]
- Redondo, R.L.; Morris, R.G. Making memories last: The synaptic tagging and capture hypothesis. Nat. Rev. Neurosci. 2011, 12, 17–30. [Google Scholar] [CrossRef]
- Nguyen, P.V.; Woo, N.H. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog. Neurobiol. 2003, 71, 401–437. [Google Scholar] [CrossRef]
- Bourtchuladze, R.; Frenguelli, B.; Blendy, J.; Cioffi, D.; Schutz, G.; Silva, A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 1994, 79, 59–68. [Google Scholar] [CrossRef]
- Abel, T.; Nguyen, P.V.; Barad, M.; Deuel, T.A.; Kandel, E.R.; Bourtchouladze, R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell 1997, 88, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Moya, S.M.; Bauer, K.E.; Kiebler, M.A. Meet the players: Local translation at the synapse. Front. Mol. Neurosci. 2014, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Yasuda, M.; Coats, J.K.; Jones, Y.; Martone, M.E.; Mayford, M. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 2002, 36, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Farris, S.; Ward, J.M.; Carstens, K.E.; Samadi, M.; Wang, Y.; Dudek, S.M. Hippocampal Subregions Express Distinct Dendritic Transcriptomes that Reveal Differences in Mitochondrial Function in CA2. Cell Rep. 2019, 29, 522–539.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingi, T.; Krumins, A.M.; Chidiac, P.; Brothers, G.M.; Chung, S.; Snow, B.E.; Barnes, C.A.; Lanahan, A.A.; Siderovski, D.P.; Ross, E.M.; et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J. Neurosci. 1998, 18, 7178–7188. [Google Scholar] [CrossRef] [PubMed]
- English, J.D.; Sweatt, J.D. A requirement for the mitogen-activated protein kinase cascade in hippocampal long-term potentiation. J. Biol. Chem. 1997, 272, 19103–19106. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Storm, D.R. The role of calmodulin as a signal integrator for synaptic plasticity. Nat. Rev. Neurosci. 2005, 6, 267–276. [Google Scholar] [CrossRef]
- Ferguson, G.D.; Storm, D.R. Why calcium-stimulated adenylyl cyclases? Physiology 2004, 19, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Visel, A.; Alvarez-Bolado, G.; Thaller, C.; Eichele, G. Comprehensive analysis of the expression patterns of the adenylate cyclase gene family in the developing and adult mouse brain. J. Comp. Neurol. 2006, 496, 684–697. [Google Scholar] [CrossRef]
- Wong, S.T.; Athos, J.; Figueroa, X.A.; Pineda, V.V.; Schaefer, M.L.; Chavkin, C.C.; Muglia, L.J.; Storm, D.R. Calcium-stimulated adenylyl cyclase activity is critical for hippocampus-dependent long-term memory and late phase LTP. Neuron 1999, 23, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Makhinson, M.; Chotiner, J.K.; Watson, J.B.; O’Dell, T.J. Adenylyl cyclase activation modulates activity-dependent changes in synaptic strength and Ca2+/calmodulin-dependent kinase II autophosphorylation. J. Neurosci. 1999, 19, 2500–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stornetta, R.L.; Zhu, J.J. Ras and Rap signaling in synaptic plasticity and mental disorders. Neuroscientist 2011, 17, 54–78. [Google Scholar] [CrossRef]
- Zhu, J.J.; Qin, Y.; Zhao, M.; Van Aelst, L.; Malinow, R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 2002, 110, 443–455. [Google Scholar] [CrossRef] [Green Version]
- Harvey, C.D.; Yasuda, R.; Zhong, H.; Svoboda, K. The spread of Ras activity triggered by activation of a single dendritic spine. Science 2008, 321, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.T.; Hsieh, T.Y.; Tsai, T.C.; Chen, C.C.; Huang, C.C.; Hsu, K.S. Conditional Deletion of Hippocampal CA2/CA3a Oxytocin Receptors Impairs the Persistence of Long-Term Social Recognition Memory in Mice. J. Neurosci. 2018, 38, 1218–1231. [Google Scholar] [CrossRef] [Green Version]
- Pagani, J.H.; Zhao, M.; Cui, Z.; Avram, S.K.; Caruana, D.A.; Dudek, S.M.; Young, W.S. Role of the vasopressin 1b receptor in rodent aggressive behavior and synaptic plasticity in hippocampal area CA2. Mol. Psychiatry 2015, 20, 490–499. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, A.; Baby, N.; Krishna, K.; Hakim, M.; Wong, Y.P.; Behnisch, T.; Soong, T.W.; Sajikumar, S. Substance P induces plasticity and synaptic tagging/capture in rat hippocampal area CA2. Proc. Natl. Acad. Sci. USA 2017, 114, E8741–E8749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, S.B.; Caruana, D.A.; Zhao, M.; Dudek, S.M. Caffeine-induced synaptic potentiation in hippocampal CA2 neurons. Nat. Neurosci. 2011, 15, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Munoz, M.D.; Solis, J.M. Characterisation of the mechanisms underlying the special sensitivity of the CA2 hippocampal area to adenosine receptor antagonists. Neuropharmacology 2019, 144, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hollinger, S.; Ramineni, S.; Hepler, J.R. Phosphorylation of RGS14 by protein kinase A potentiates its activity toward Gαi. Biochemistry 2003, 42, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Hollinger, S.; Hepler, J.R. Methods for measuring RGS protein phosphorylation by G protein-regulated kinases. Methods Mol. Biol. 2004, 237, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Watanabe, M.; Offermanns, S.; Simon, M.I.; Kano, M. Group I metabotropic glutamate receptor signaling via Gαq/Gα11 secures the induction of long-term potentiation in the hippocampal area CA1. J. Neurosci. 2002, 22, 8379–8390. [Google Scholar] [CrossRef] [Green Version]
- Boehm, J.; Kang, M.G.; Johnson, R.C.; Esteban, J.; Huganir, R.L.; Malinow, R. Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron 2006, 51, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Williams Avram, S.K.; Lee, H.J.; Fastman, J.; Cymerblit-Sabba, A.; Smith, A.; Vincent, M.; Song, J.; Granovetter, M.C.; Lee, S.H.; Cilz, N.I.; et al. NMDA Receptor in Vasopressin 1b Neurons Is Not Required for Short-Term Social Memory, Object Memory or Aggression. Front. Behav. Neurosci. 2019, 13, 218. [Google Scholar] [CrossRef] [Green Version]
- Broadbent, N.J.; Gaskin, S.; Squire, L.R.; Clark, R.E. Object recognition memory and the rodent hippocampus. Learn. Mem. 2010, 17, 5–11. [Google Scholar] [CrossRef] [Green Version]
- Broadbent, N.J.; Squire, L.R.; Clark, R.E. Spatial memory, recognition memory, and the hippocampus. Proc. Natl. Acad. Sci. USA 2004, 101, 14515–14520. [Google Scholar] [CrossRef] [Green Version]
- Alexander, G.M.; Riddick, N.V.; McCann, K.E.; Lustberg, D.; Moy, S.S.; Dudek, S.M. Modulation of CA2 neuronal activity increases behavioral responses to fear conditioning in female mice. Neurobiol. Learn. Mem. 2019, 163, 107044. [Google Scholar] [CrossRef]
- Marschner, A.; Kalisch, R.; Vervliet, B.; Vansteenwegen, D.; Buchel, C. Dissociable roles for the hippocampus and the amygdala in human cued versus context fear conditioning. J. Neurosci. 2008, 28, 9030–9036. [Google Scholar] [CrossRef] [Green Version]
- Masmudi-Martin, M.; Navarro-Lobato, I.; López-Aranda, M.F.; Delgado, G.; Martin-Montañez, E.; Quiros-Ortega, M.E.; Carretero-Rey, M.; Narváez, L.; Garcia-Garrido, M.F.; Posadas, S.; et al. RGS14414 treatment induces memory enhancement and rescues episodic memory deficits. FASEB J. 2019, 33, 11804–11820. [Google Scholar] [CrossRef] [Green Version]
- López-Aranda, M.F.; López-Téllez, J.F.; Navarro-Lobato, I.; Masmudi-Martín, M.; Gutiérrez, A.; Khan, Z.U. Role of layer 6 of V2 visual cortex in object-recognition memory. Science 2009, 325, 87–89. [Google Scholar] [CrossRef] [Green Version]
- LaLumiere, R.T.; McGaugh, J.L.; McIntyre, C.K. Emotional Modulation of Learning and Memory: Pharmacological Implications. Pharmacol. Rev. 2017, 69, 236–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, C.C.; Sokoloff, G.; Cheng, R.; Palmer, A.A. Genome-wide association for fear conditioning in an advanced intercross mouse line. Behav. Genet. 2012, 42, 437–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikemoto, S.; Panksepp, J. The role of nucleus accumbens dopamine in motivated behavior: A unifying interpretation with special reference to reward-seeking. Brain Res. Rev. 1999, 31, 6–41. [Google Scholar] [CrossRef]
- Ikemoto, S. Ventral striatal anatomy of locomotor activity induced by cocaine, D-amphetamine, dopamine and D1/D2 agonists. Neuroscience 2002, 113, 939–955. [Google Scholar] [CrossRef]
- Chakravarthy, V.S.; Joseph, D.; Bapi, R.S. What do the basal ganglia do? A modeling perspective. Biol. Cybern. 2010, 103, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Stocco, A.; Lebiere, C.; Anderson, J.R. Conditional routing of information to the cortex: A model of the basal ganglia’s role in cognitive coordination. Psychol. Rev. 2010, 117, 541–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pape, H.C.; Pare, D. Plastic synaptic networks of the amygdala for the acquisition, expression, and extinction of conditioned fear. Physiol. Rev. 2010, 90, 419–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haber, S.N. The place of dopamine in the cortico-basal ganglia circuit. Neuroscience 2014, 282, 248–257. [Google Scholar] [CrossRef] [Green Version]
- McNamara, C.G.; Dupret, D. Two sources of dopamine for the hippocampus. Trends Neurosci. 2017, 40, 383–384. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Kochubey, O.; Kintscher, M.; Schneggenburger, R. A VTA to Basal Amygdala Dopamine Projection Contributes to Signal Salient Somatosensory Events during Fear Learning. J. Neurosci. 2020, 40, 3969–3980. [Google Scholar] [CrossRef]
- Asl, M.M.; Vahabie, A.H.; Valizadeh, A. Dopaminergic Modulation of Synaptic Plasticity, Its Role in Neuropsychiatric Disorders, and Its Computational Modeling. Basic Clin. Neurosci. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Ghavami, A.; Hunt, R.A.; Olsen, M.A.; Zhang, J.; Smith, D.L.; Kalgaonkar, S.; Rahman, Z.; Young, K.H. Differential effects of regulator of G protein signaling (RGS) proteins on serotonin 5-HT1A, 5-HT2A, and dopamine D2 receptor-mediated signaling and adenylyl cyclase activity. Cell. Signal. 2004, 16, 711–721. [Google Scholar] [CrossRef]
- Gross, J.D.; Kaski, S.W.; Schroer, A.B.; Wix, K.A.; Siderovski, D.P.; Setola, V. Regulator of G protein signaling-12 modulates the dopamine transporter in ventral striatum and locomotor responses to psychostimulants. J. Psychopharmacol. 2018, 32, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Lerner, T.N.; Kreitzer, A.C. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron 2012, 73, 347–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, Z.; Schwarz, J.; Gold, S.J.; Zachariou, V.; Wein, M.N.; Choi, K.H.; Kovoor, A.; Chen, C.K.; DiLeone, R.J.; Schwarz, S.C.; et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron 2003, 38, 941–952. [Google Scholar] [CrossRef] [Green Version]
- Stewart, A.; Maity, B.; Anderegg, S.P.; Allamargot, C.; Yang, J.; Fisher, R.A. Regulator of G protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc. Natl. Acad. Sci. USA 2015, 112, E786–E795. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Guzman, J.N.; Tkatch, T.; Chen, S.; Goldberg, J.A.; Ebert, P.J.; Levitt, P.; Wilson, C.J.; Hamm, H.E.; Surmeier, D.J. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat. Neurosci. 2006, 9, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Omer, H. Specifics and nonspecifics in psychotherapy. Am. J. Psychother. 1989, 43, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Mark, M.D.; Li, X.; Xie, M.; Waka, S.; Rettig, J.; Herlitze, S. RGS2 determines short-term synaptic plasticity in hippocampal neurons by regulating Gi/o-mediated inhibition of presynaptic Ca2+ channels. Neuron 2006, 51, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Besnard, A.; Bouveyron, N.; Kappes, V.; Pascoli, V.; Pages, C.; Heck, N.; Vanhoutte, P.; Caboche, J. Alterations of molecular and behavioral responses to cocaine by selective inhibition of Elk-1 phosphorylation. J. Neurosci. 2011, 31, 14296–14307. [Google Scholar] [CrossRef]
- Britt, J.P.; Benaliouad, F.; McDevitt, R.A.; Stuber, G.D.; Wise, R.A.; Bonci, A. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron 2012, 76, 790–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourrich, S.; Rothwell, P.E.; Klug, J.R.; Thomas, M.J. Cocaine experience controls bidirectional synaptic plasticity in the nucleus accumbens. J. Neurosci. 2007, 27, 7921–7928. [Google Scholar] [CrossRef] [Green Version]
- Belin, D.; Jonkman, S.; Dickinson, A.; Robbins, T.W.; Everitt, B.J. Parallel and interactive learning processes within the basal ganglia: Relevance for the understanding of addiction. Behav. Brain Res. 2009, 199, 89–102. [Google Scholar] [CrossRef]
- Belin, D.; Everitt, B.J. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron 2008, 57, 432–441. [Google Scholar] [CrossRef] [Green Version]
- Werner, C.T.; Mitra, S.; Auerbach, B.D.; Wang, Z.J.; Martin, J.A.; Stewart, A.F.; Gobira, P.H.; Iida, M.; An, C.; Cobb, M.M.; et al. Neuroadaptations in the dorsal hippocampus underlie cocaine seeking during prolonged abstinence. Proc. Natl. Acad. Sci. USA 2020, 117, 26460–26469. [Google Scholar] [CrossRef]
- Squires, K.E.; Montañez-Miranda, C.; Pandya, R.R.; Torres, M.P.; Hepler, J.R. Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease. Pharmacol. Rev. 2018, 70, 446–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazer, K.A.; Murray, S.S.; Schork, N.J.; Topol, E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009, 10, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Weinman, E.J.; Hall, R.A.; Friedman, P.A.; Liu-Chen, L.Y.; Shenolikar, S. The association of NHERF adaptor proteins with G protein-coupled receptors and receptor tyrosine kinases. Annu. Rev. Physiol. 2006, 68, 491–505. [Google Scholar] [CrossRef] [Green Version]
- Iasevoli, F.; Tomasetti, C.; de Bartolomeis, A. Scaffolding proteins of the post-synaptic density contribute to synaptic plasticity by regulating receptor localization and distribution: Relevance for neuropsychiatric diseases. Neurochem. Res. 2013, 38, 1–22. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harbin, N.H.; Bramlett, S.N.; Montanez-Miranda, C.; Terzioglu, G.; Hepler, J.R. RGS14 Regulation of Post-Synaptic Signaling and Spine Plasticity in Brain. Int. J. Mol. Sci. 2021, 22, 6823. https://doi.org/10.3390/ijms22136823
Harbin NH, Bramlett SN, Montanez-Miranda C, Terzioglu G, Hepler JR. RGS14 Regulation of Post-Synaptic Signaling and Spine Plasticity in Brain. International Journal of Molecular Sciences. 2021; 22(13):6823. https://doi.org/10.3390/ijms22136823
Chicago/Turabian StyleHarbin, Nicholas H., Sara N. Bramlett, Carolina Montanez-Miranda, Gizem Terzioglu, and John R. Hepler. 2021. "RGS14 Regulation of Post-Synaptic Signaling and Spine Plasticity in Brain" International Journal of Molecular Sciences 22, no. 13: 6823. https://doi.org/10.3390/ijms22136823