Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails
Abstract
:1. Introduction
2. Biocatalysts for Cellulose Degradation
3. Advantages of the Enzymatic Complex of Filamentous Fungi Penicillium
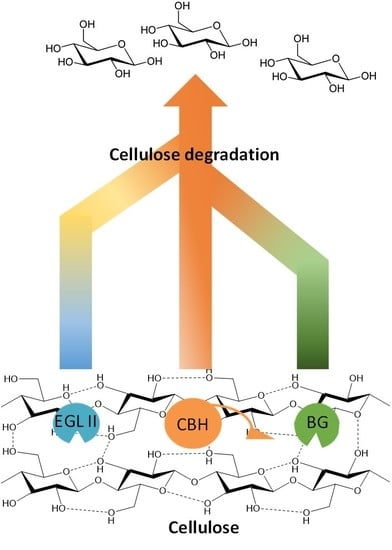
4. Cellulases Synergism
5. Cellulolytic Cocktails
| Cocktails | Composition (Type of Enzyme) | Substrate | Effect | Reference |
|---|---|---|---|---|
| Artificial cocktails of purified cellulases from Chrysosporium lucknowense and Trichoderma reesei | CBH Ia, Ib, and IIb; endoglucanases II and V; β-glucosidase, xylanase II | Pretreated Douglas fir wood, cotton, 1MCC | Efficient saccharification | [36] |
| Optimization of cellulases, accessory enzymes and additives in high-solids hydrolysates | Cellulases and accessory enzymes | Pretreated sugarcane bagasse | High yield of simple sugars | [42] |
| Cellulase and hemicellulase synergy | a-L-arabino-furanosidase, xylanase, and cellulases | Pretreated cornstalk and corn bran | High yield of simple sugars | [39] |
| Cellobiohydrolases, endoglucanases, and β-glucosidase from Talaromyces cellulolyticus | Cel5A, Cel6A, Cel7A, Cel7B, and Xyl10A, Bgl3A | Acid-pretreated corn stover, Avicel | Lower enzyme load | [40] |
| Penicillium verruculosum cellulase cocktail B1-537 | CBH I, EG II P. verruculosum | 1MCC, Aspen | 28% conversion on Aspen wood after 24 h at 50 °C pH 5.0 | [38] |
| Penicillium verruculosum cellulase cocktail BGL + EG IV | CBH I, EG II P. verruculosum, BGL A. niger, and EGIV T. reesei | 1MCC, Aspen | 35% conversion on Aspen wood after 24 h at 50 °C pH 5.0 | [38] |
| Penicillium verruculosum cellulase cocktail CBH I+EG II+BGL | CBH I, EG II P. verruculosum and BGL A. niger | 1MCC, Aspen | 30% conversion on Aspen wood after 24 h at 50 °C pH 5.0 | [38] |
| Penicillium verruculosum cellulase cocktail B1-151 + F10 | Cellulases P. verruculosum and BGL A. niger | 1MCC, Aspen | 45% conversion on Aspen wood after 24 h at 50 °C pH 5.0 | [37,38] |
| Accelerase 1500 | CBH, EG T. reesei, BGL A. niger and other | 1MCC, Aspen | 35% conversion on Aspen wood after 48 h at 50 °C pH 5.0 | [37] |
| Cellic Ctec-1 | CBH, EG T. reesei, BGL A. oryzae and other | 1MCC, Aspen | 19% conversion on Aspen wood after 48 h at 50 °C pH 5.0 | [37] |
| Cellic Ctec-2 | CBH, EG T. reesei, BGL A. fumigatus and other | 1MCC, Aspen | 40% conversion on Aspen wood after 48 h at 50 °C pH 5.0 | [37] |
Challenges of Cellulases Cocktails
6. Protein Engineering for Tailored Cellulases Cocktails
6.1. Engineering Cellulases for Enhanced Activity for Cellulose Degradation
6.2. Engineering Cellulases for Enhanced Thermostability
| Cellulase (Source) | Improvement | Engineering Method | Activity Assay | Molecular Effect | Reference |
|---|---|---|---|---|---|
| β-glucosidase A (Clostridium thermocellum) | 6.4 °C in Tm (from 79.3 to 85.7 °C) | Directed evolution— error-prone PCR | 4pNPG | N.A. | [120] |
| Endoglucanase PvCel5A (Penicillium verruculosum) | Increase in half-life activity by 1.5-2-fold at 70 and 80 °C | Rational design—Disulfide bond engineering | 10CMC/ 11NS β-glucan/ 11NS | increase the overall compactness of the structure | [121] |
| Endoglucanase TeEgl5A (Talaromyces emersonii) | Increase of Tm by 10 °C and 1.6-fold improvement of specific activity | Semi-Rational design—SCHEMA | 10CMC/ 6DNS | improved hydrophobic packing | [122] |
| Cellobiohydrolase CBH I (Talaromyces cellulolyticus) | Increase in 8.0 °C in Tm | Rational design—DNA shuffling with homologous enzymes | 1pNPL | Increased interaction stabilizes protein | [79] |
| β-glucosidase Ks5A7 (GeneBank: HV348683) | 25.5 °C improvement in T50 | Directed evolution—ep-PCR | 8GOD–POD assay kit | N.A. | [123] |
| Cellobiohydrolase Cel7A (Rasamsonia emersonii) | Acceleration by temperature (about two-fold faster around 70 °C) | Semi-rational—CBD fusion | Cellulose/ 3PAHBAH | N.A. | [14] |
| Cellobiohydrolase Cel7A (Trichoderma reesei) | 10.4 °C increase in Tm | Semi-rational design—MSA with thermostable homologs and DNA shuffling | 94-MUC | Strengthen of hydrophobic interactions | [116] |
| Endoglucanase Z (Clostridium cellulovorans) | Optimal temperature increased by 7.5 °C | Rational design—Amino acid alignment with thermostable cellulases | 10CMC/ 6DNS | stabilization of the active site and the improvement of the rigid folding structure | [97] |
| Endoglucanase Cel9A (Alicyclobacillus acidocaldarius) | 5.9 °C increase in Tm | Rational design—Engineering a calcium-binding residue | 10CMC/ 6DNS | Stabilization of Ca binding site | [101] |
| Endoglucanase GsCelA and BsCel5A (Geobacillus sp. and Bacillus sp.) | Increase of T50 by 4 °C | Semi-rational design—SCHEMA | 7PASC/ 6DNS | Increase hydrophobic amino acid in the buried protein environment | [115] |
| Endoglucanase I (Trichoderma reesei) | 25% increase in thermal stability at 65 °C for 8 h | Rational design—SDM based in free energy stabilization and MD | Azo-10CMC 10CMC and 7PASC/ 6DNS | Thermodynamic stabilization | [102] |
| Endoglucanase Cel12B (Thermotoga maritima) | Retain 90% of activity after 8 h at 80 °C | Rational design—Homology modeling | 10CMC/ 6DNS | Increase the hydrophobicity of the outer surface to form a more compact complex with the substrate | [103] |
| Endoglucanase I (Trichoderma reesei) | Increase Tm in 3 °C and t1/2 at 60 °C of 80 h | Rational design—B-factor guided approach | 10CMC/ 6DNS | Rigidification of mobile portions of the structure | [104] |
| α-glucosidase TtAG (Thermus thermophilus) | 4 °C improvement in T50 (97 °C) | Directed evolution—ep-PCR | 4pNPG | N.A. | [124] |
| Cellobiohydrolase Cel7A (Talaromyces emersonii) | Increase the Tm by 4 °C | Rational design—CBD fusion and disulfide bridge in the catalytic site | 124-MUL and 13CNPLac | N.A. | [107] |
| Endoglucanase Cel7B (Hypocrea pseudokoningii) | Increase of T50 in 7 °C | Semi-rational design—Random mutagenesis, comparison with homolog and cavity stabilization | 124-MUL | N.A. | [125] |
| Endoglucanase Cel5A (GeneBank: JN012243) | 7-fold increase in thermostability at 65 °C | Directed evolution—ep-PCR and CBD fusion | 10CMC/ 6DNS | Increase compactness and stability around the active site | [106] |
| Cellobiohydrolase CBH II (Phanerochaete chrysosporium) | Increase of T50120 by 5.4 °C | Semi-rational design—Consensus mutations from thermophilic cellulases | 7PASC/ 14TZ | Increase hydrophobic amino acid in the buried protein environment | [98] |
| Cellobiohydrolase Cel6A HJPlus (Chimera) | Variant 3C6P has a t1/2 of 280 min at 75 °C and a T50 of 80.1 °C | Directed evolution—ep-PCR | 2MCC/ 11NS | Increase in hydrophobicity and limited conformational freedom due to proline substitutions | [126] |
| Cellobiohydrolase Cel7A (Trichoderma reesei) | Increase of T50 by 3 °C | Semi-rational design—non continuous recombination | 124-MUL | Rigidification by introduction of a hydrogen bond | [127] |
| Endoglucanase celC (Clostridium thermocellum) | Increase the Tm by 4 °C | Rational design—SDM (stabilizing positions) and disulfide bond formation retrieved from MD | 5pNPC | Improvement of local protein stability | [105] |
| Cellobiohydrolase Cel7A (Talaromyces emersonii) | Improved thermostability at 65 °C | Semi-rational design—Biased clique shuffling | 2MCC/ Amplex Red and 124-MUL | N.A. | [114] |
| Endoglucanase Cel8A (Clostridium thermocellum) | Increase of half-life activity by 14-fold at 85 °C | Rational design—consensus mutations from homologous GH8 | 10CMC/ 6DNS | Increased rigidity | [100] |
| Cellobiohydrolase CBH II (Chaetomium thermophilum) | More than 50% of activity after 60 min incubation at 80 °C | Directed evolution—ep-PCR | 5pNPC | Increased hydrogen bonds | [128] |
| Endoglucanase Cel5A (Clostridium phytophermentans) | 92%, 36%, and 46% longer t1/2 at 60 °C on CMC, cellulose, and MCC, respectively | Directed evolution—ep-PCR and CBD fusion | 10CMC, 2MCC and cellulose | N.A. | [112] |
| Endoglucanase Cel8A (Clostridium thermocellum) | Increase the Tm by 7.0 °C and the t1/2 by 8-fold at 85 °C. | Directed evolution—ep-PCR and shuffle | 10CMC/ 6DNS | N.A. | [113] |
| Cellobiohydrolase CBH I (Talaromyces emersonii) | Increase of T50 by 3.4 °C | Semi-rational—SCHEMA | 124-MUL | N.A. | [129] |
| Cellobiohydrolase TeCel7A (Talaromyces emersonii) | Increase of Tm by 9 °C | Rational design—Introduction of disulfide bonds | 124-MUL | Rigidification by introduction of disulfide bond | [130] |
6.3. Engineering Cellulases for Enhanced Performance in Non-conventional Media (Ionic Liquids, High Salt Concentration, Organic Solvents)
6.4. Engineering Cellulase for pH Stability
6.5. Robust Cellulases for Cellulolytic Cocktails
7. Future Perspective of Tailored Cellulases Cocktails
Author Contributions
Funding
Conflicts of Interest
References
- Fischer, G.; Prieler, S.; van Velthuizen, H.; Lensink, S.M.; Londo, M.; de Wit, M. Biofuel production potentials in Europe: Sustainable use of cultivated land and pastures. Part I: Land productivity potentials. Biomass Bioenergy 2010, 34, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Quiroz-Castañeda, R.E.; Folch-Mallol, J.L. Hydrolysis of biomass mediated by cellulases for the production of sugars. In Sustainable Degradation of Lignocellulosic Biomass—Techniques, Applications and Commercialization; InTech: London, UK, 2013; pp. 119–155. [Google Scholar] [CrossRef]
- Tabil, L.; Adapa, P.; Kashaninejad, M. Biomass feedstock pre-processing—Part 1: Pre-treatment. In Biofuel’s Engineering Process Technology; InTech: London, UK, 2011. [Google Scholar]
- Bhattacharya, A.S.; Bhattacharya, A.; Pletschke, B.I. Synergism of fungal and bacterial cellulases and hemicellulases: A novel perspective for enhanced bio-ethanol production. Biotechnol. Lett. 2015, 37, 1117–1129. [Google Scholar] [CrossRef]
- Lopes, A.; Ferreira Filho, E.; Moreira, L. An update on enzymatic cocktails for lignocellulose breakdown. J. Appl. Microbiol. 2018, 125, 632–645. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Singh, S.; Singh, O.V. Bioconversion of lignocellulosic biomass: Biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 2008, 35, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Z.; Fang, X.; Wang, L.; Qu, Y. Cellulolytic enzyme production and enzymatic hydrolysis for second-generation bioethanol production. In Biotechnology in China III: Biofuels and Bioenergy; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–24. [Google Scholar]
- Pérez, J.; Munoz-Dorado, J.; De la Rubia, T.; Martinez, J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: An overview. Int. Microbiol. 2002, 5, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, T.; Karlsson, J.; Tjerneld, F. A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (Cel7A) and endoglucanase I (Cel7B) of Trichoderma reesei. Appl. Biochem. Biotechnol. 2002, 101, 41–60. [Google Scholar] [CrossRef]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Sukharnikov, L.O.; Cantwell, B.J.; Podar, M.; Zhulin, I.B. Cellulases: Ambiguous nonhomologous enzymes in a genomic perspective. Trends Biotechnol. 2011, 29, 473–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badino, S.F.; Christensen, S.J.; Kari, J.; Windahl, M.S.; Hvidt, S.; Borch, K.; Westh, P. Exo-exo synergy between Cel6A and Cel7A from Hypocrea jecorina: Role of carbohydrate binding module and the endo-lytic character of the enzymes. Biotechnol. Bioeng. 2017, 114, 1639–1647. [Google Scholar] [CrossRef] [Green Version]
- Westh, P.; Borch, K.; Sørensen, T.; Tokin, R.; Kari, J.; Badino, S.; Cavaleiro, M.A.; Røjel, N.; Christensen, S.; Vesterager, C.S. Thermoactivation of a cellobiohydrolase. Biotechnol. Bioeng. 2018, 115, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Heitmann, J.A., Jr.; Rojas, O.J. Quantification of cellulase activity using the quartz crystal microbalance technique. Anal. Chem. 2009, 81, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Selig, M.J.; Vuong, T.V.; Gudmundsson, M.; Forsberg, Z.; Westereng, B.; Felby, C.; Master, E.R. Modified cellobiohydrolase–cellulose interactions following treatment with lytic polysaccharide monooxygenase CelS2 (ScLPMO10C) observed by QCM-D. Cellulose 2015, 22, 2263–2270. [Google Scholar] [CrossRef]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Stahlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gusakov, A.V. Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 2011, 29, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Gusakov, A.V.; Sinitsyn, A.P. Cellulases from Penicillium species for producing fuels from biomass. Biofuels 2012, 3, 463–477. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, L.; Wei, X.; Zou, G.; Qin, Y.; Ma, L.; Li, J.; Zheng, H.; Wang, S.; Wang, C. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 2013, 8, e55185. [Google Scholar] [CrossRef] [Green Version]
- Ogunmolu, F.E.; Kaur, I.; Gupta, M.; Bashir, Z.; Pasari, N.; Yazdani, S.S. Proteomics insights into the biomass hydrolysis potentials of a hypercellulolytic fungus Penicillium funiculosum. J. Proteome Res. 2015, 14, 4342–4358. [Google Scholar] [CrossRef]
- Hu, L.; Taujale, R.; Liu, F.; Song, J.; Yin, Q.; Zhang, Y.; Guo, J.; Yin, Y. Draft genome sequence of Talaromyces verruculosus (“Penicillium verruculosum”) strain TS63-9, a fungus with great potential for industrial production of polysaccharide-degrading enzymes. J. Biotechnol. 2016, 219, 5–6. [Google Scholar] [CrossRef]
- Morozova, V.V.; Gusakov, A.V.; Andrianov, R.M.; Pravilnikov, A.G.; Osipov, D.O.; Sinitsyn, A.P. Cellulases of Penicillium verruculosum. Biotechnol. J. 2010, 5, 871–880. [Google Scholar] [CrossRef]
- Volkov, P.V.; Rozhkova, A.M.; Gusakov, A.V.; Sinitsyn, A.P. Homologous cloning, purification and characterization of highly active cellobiohydrolase I (Cel7A) from Penicillium canescens. Protein Expr. Purif. 2014, 103, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dotsenko, A.S.; Gusakov, A.V.; Rozhkova, A.M.; Sinitsyna, O.A.; Nemashkalov, V.A.; Sinitsyn, A.P. Effect of N-linked glycosylation on the activity and other properties of recombinant endoglucanase IIa (Cel5A) from Penicillium verruculosum. Protein Eng. Des. Sel. 2016, 29, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Dotsenko, A.S.; Gusakov, A.V.; Volkov, P.V.; Rozhkova, A.M.; Sinitsyn, A.P. N-linked glycosylation of recombinant cellobiohydrolase I (Cel7A) from Penicillium verruculosum and its effect on the enzyme activity. Biotechnol. Bioeng. 2016, 113, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Thoresen, M.; van Dyk, J.S.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzym. Microb. Technol. 2017, 103, 1–11. [Google Scholar] [CrossRef]
- Dotsenko, A.; Gusakov, A.; Rozhkova, A.; Sinitsyna, O.; Shashkov, I.; Sinitsyn, A. Enzymatic hydrolysis of cellulosic materials using synthetic mixtures of purified cellulases bioengineered at N-glycosylation sites. 3 Biotech 2018, 8, 396. [Google Scholar] [CrossRef]
- Rahikainen, J.; Ceccherini, S.; Molinier, M.; Holopainen-Mantila, U.; Reza, M.; Väisänen, S.; Puranen, T.; Kruus, K.; Vuorinen, T.; Maloney, T. Effect of cellulase family and structure on modification of wood fibres at high consistency. Cellulose 2019, 26, 5085–5103. [Google Scholar] [CrossRef] [Green Version]
- Boisset, C.; Pétrequin, C.; Chanzy, H.; Henrissat, B.; Schülein, M. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 2001, 72, 339–345. [Google Scholar] [CrossRef]
- Jalak, J.; Kurašin, M.; Teugjas, H.; Väljamäe, P. Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 2012, 287, 28802–28815. [Google Scholar] [CrossRef] [Green Version]
- Jeoh, T.; Wilson, D.B.; Walker, L.P. Effect of cellulase mole fraction and cellulose recalcitrance on synergism in cellulose hydrolysis and binding. Biotechnol. Prog. 2006, 22, 270–277. [Google Scholar] [CrossRef]
- Sukumaran, R.K.; Singhania, R.R.; Mathew, G.M.; Pandey, A. Cellulase production using biomass feed stock and its application in lignocellulose saccharification for bio-ethanol production. Renew. Energy 2009, 34, 421–424. [Google Scholar] [CrossRef]
- Singhania, R.R. Production of celluloytic enzymes for the hydrolysis of lignocellulosic biomass. In Biofuels; Elsevier: Amsterdam, The Netherlands, 2011; pp. 177–201. [Google Scholar]
- Teter, S.A.; Sutton, K.B.; Emme, B. Enzymatic processes and enzyme development in biorefining. In Advances in Biorefineries; Elsevier: Amsterdam, The Netherlands, 2014; pp. 199–233. [Google Scholar]
- Gusakov, A.V.; Salanovich, T.N.; Antonov, A.I.; Ustinov, B.B.; Okunev, O.N.; Burlingame, R.; Emalfarb, M.; Baez, M.; Sinitsyn, A.P. Design of highly efficient cellulase mixtures for enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2007, 97, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Chekushina, A.; Dotsenko, G.; Sinitsyn, A. Comparing the efficiency of plant material bioconversion processes using biocatalysts based on Trichoderma and Penicillium verruculosum enzyme preparations. Catal. Ind. 2013, 5, 98–104. [Google Scholar] [CrossRef]
- Sinitsyn, A.; Korotkova, O.; Sinitsyna, O.; Rozhkova, A.; Dotsenko, G.; Proskurina, O.; Osipov, D.; Kondrat’eva, E.; Chekushina, A. Optimizing the composition of cellulase enzyme complex from Penicillium verruculosum: Enhancing hydrolytic capabilities via genetic engineering. Catal. Ind. 2016, 8, 101–106. [Google Scholar] [CrossRef]
- Tu, T.; Li, X.; Meng, K.; Bai, Y.; Wang, Y.; Wang, Z.; Yao, B.; Luo, H. A GH51 α-l-arabinofuranosidase from Talaromyces leycettanus strain JCM12802 that selectively drives synergistic lignocellulose hydrolysis. Microb. Cell Fact. 2019, 18, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, H.; Decker, S.R.; Taylor, L.E.; Yano, S.; Sawayama, S. Identification and characterization of core cellulolytic enzymes from Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) critical for hydrolysis of lignocellulosic biomass. Biotechnol. Biofuels 2014, 7, 151. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, R.; Semwal, S.; Kumar, R.; Mathur, A.; Gupta, R.P.; Tuli, D.K.; Satlewal, A. Synergistic enzyme eocktail to enhance hydrolysis of steam exploded wheat straw at pilot scale. Front. Energy Res. 2018, 6, 122. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, J.; Zhang, Y.; Guo, Y.; Xu, H.; Xu, J.; Wang, Z. Enhancement of high-solids enzymatic hydrolysis efficiency of alkali pretreated sugarcane bagasse at low cellulase dosage by fed-batch strategy based on optimized accessory enzymes and additives. Bioresour. Technol. 2019, 292, 121993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, J.; Luo, L.; Wang, E.; Wang, R.; Liu, L.; Liu, J.; Yuan, H. Low-Cost Cellulase-Hemicellulase Mixture Secreted by Trichoderma harzianum EM0925 with Complete Saccharification Efficacy of Lignocellulose. Int. J. Mol. Sci. 2020, 21, 371. [Google Scholar] [CrossRef] [Green Version]
- Bunterngsook, B.; Laothanachareon, T.; Chotirotsukon, C.; Inoue, H.; Fujii, T.; Hoshino, T.; Roongsawang, N.; Kuboon, S.; Kraithong, W.; Techanan, W. Development of tailor-made synergistic cellulolytic enzyme system for saccharification of steam exploded sugarcane bagasse. J. Biosci. Bioeng. 2018, 125, 390–396. [Google Scholar] [CrossRef]
- Du, J.; Liang, J.; Gao, X.; Liu, G.; Qu, Y. Optimization of an artificial cellulase cocktail for high-solids enzymatic hydrolysis of cellulosic materials with different pretreatment methods. Bioresour. Technol. 2020, 295, 122272. [Google Scholar] [CrossRef]
- Adsul, M.; Sandhu, S.K.; Singhania, R.R.; Gupta, R.; Puri, S.K.; Mathur, A. Designing a cellulolytic enzyme cocktail for the efficient and economical conversion of lignocellulosic biomass to biofuels. Enzym. Microb. Technol. 2019, 133, 109442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Pei, Z.; Wang, D. Organic solvent pretreatment of lignocellulosic biomass for biofuels and biochemicals: A review. Bioresour. Technol. 2016, 199, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Galbe, M.; Wallberg, O. Pretreatment for biorefineries: A review of common methods for efficient utilisation of lignocellulosic materials. Biotechnol. Biofuels 2019, 12, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Lee, Y.; Kim, T.H. A review on alkaline pretreatment technology for bioconversion of lignocellulosic biomass. Bioresour. Technol. 2016, 199, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.K.; Vyas, P.; Dubey, A.; Upadhyaya, C.P.; Kothari, R.; Tyagi, V.; Kumar, A. Assessment of different pretreatment technologies for efficient bioconversion of lignocellulose to ethanol. Front. Biosci. Sch. Ed. 2018, 10, 10–2741. [Google Scholar]
- Bhatia, S.K.; Jagtap, S.S.; Bedekar, A.A.; Bhatia, R.K.; Patel, A.K.; Pant, D.; Banu, J.R.; Rao, C.V.; Kim, Y.-G.; Yang, Y.-H. Recent developments in pretreatment technologies on lignocellulosic biomass: Effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 2020, 300, 122724. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, R.J.; Sweeney, M.D.; Leggio, L.L.; Otten, H.; Poulsen, J.-C.N.; Johansen, K.S.; Krogh, K.B.; Jørgensen, C.I.; Tovborg, M.; Anthonsen, A. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef] [Green Version]
- Frommhagen, M.; Westphal, A.H.; Van Berkel, W.J.; Kabel, M.A. Distinct substrate specificities and electron-donating systems of fungal lytic polysaccharide monooxygenases. Front. Microbiol. 2018, 9, 1080. [Google Scholar] [CrossRef]
- Santo, M.E.; Rezende, C.A.; Bernardinelli, O.D.; Pereira, N., Jr.; Curvelo, A.A.; Deazevedo, E.R.; Guimarães, F.E.; Polikarpov, I. Structural and compositional changes in sugarcane bagasse subjected to hydrothermal and organosolv pretreatments and their impacts on enzymatic hydrolysis. Ind. Crops Prod. 2018, 113, 64–74. [Google Scholar] [CrossRef]
- Kim, D. Physico-chemical conversion of lignocellulose: Inhibitor effects and detoxification strategies: A mini review. Molecules 2018, 23, 309. [Google Scholar] [CrossRef] [Green Version]
- vom Stein, T.; Grande, P.; Sibilla, F.; Commandeur, U.; Fischer, R.; Leitner, W.; de María, P.D. Salt-assisted organic-acid-catalyzed depolymerization of cellulose. Green Chem. 2010, 12, 1844–1849. [Google Scholar] [CrossRef]
- Swain, M.; Natarajan, V.; Krishnan, C. Chapter Nine-Marine Enzymes and Microorganisms for Bioethanol Production. Adv. Food Nutr. Res. 2017, 80, 181–197. [Google Scholar] [PubMed]
- Wu, M.; Mintz, M.; Wang, M.; Arora, S. Consumptive Water Use in the Production of Ethanol and Petroleum Gasoline; ESD/09-1; Argonne National Laboratory ANL: Lemont, IL, USA, 2009. Available online: www.transportation.anl.gov/pdfs/AF/557.pdf (accessed on 24 February 2020).
- Fang, C.; Thomsen, M.H.; Brudecki, G.P.; Cybulska, I.; Frankær, C.G.; Bastidas-Oyanedel, J.R.; Schmidt, J.E. Seawater as alternative to freshwater in pretreatment of date palm residues for bioethanol production in coastal and/or arid areas. ChemSusChem 2015, 8, 3823–3831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, M.L.; Puri, M.; Barrow, C.J. Recent trends in nanomaterials immobilised enzymes for biofuel production. Crit. Rev. Biotechnol. 2016, 36, 108–119. [Google Scholar] [CrossRef]
- Khoshnevisan, K.; Poorakbar, E.; Baharifar, H.; Barkhi, M. Recent advances of cellulase immobilization onto magnetic nanoparticles: An update review. Magnetochemistry 2019, 5, 36. [Google Scholar] [CrossRef] [Green Version]
- Mohanram, S.; Amat, D.; Choudhary, J.; Arora, A.; Nain, L. Novel perspectives for evolving enzyme cocktails for lignocellulose hydrolysis in biorefineries. Sustain. Chem. Process. 2013, 1, 15. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.K.; Singhania, R.R.; Sim, S.J.; Pandey, A. Thermostable cellulases: Current status and perspectives. Bioresour. Technol. 2019, 279, 385–392. [Google Scholar] [CrossRef]
- Merino, S.T.; Cherry, J. Progress and challenges in enzyme development for biomass utilization. In Biofuels; Springer: Berlin/Heidelberg, Germany, 2007; pp. 95–120. [Google Scholar]
- Olsen, J.P.; Borch, K.; Westh, P. Endo/exo-synergism of cellulases increases with substrate conversion. Biotechnol. Bioeng. 2017, 114, 696–700. [Google Scholar] [CrossRef]
- Trudeau, D.L.; Lee, T.M.; Arnold, F.H. Engineered thermostable fungal cellulases exhibit efficient synergistic cellulose hydrolysis at elevated temperatures. Biotechnol. Bioeng. 2014, 111, 2390–2397. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T.; Hauer, B.; Jaeger, K.E.; Schwaneberg, U. Directed evolution empowered redesign of natural proteins for the sustainable production of chemicals and pharmaceuticals. Angew. Chem. Int. Ed. 2019, 58, 36–40. [Google Scholar] [CrossRef]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379. [Google Scholar] [CrossRef] [PubMed]
- Shivange, A.V.; Marienhagen, J.; Mundhada, H.; Schenk, A.; Schwaneberg, U. Advances in generating functional diversity for directed protein evolution. Curr. Opin. Chem. Biol. 2009, 13, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.D.; Seelig, B. Advances in the directed evolution of proteins. Curr. Opin. Chem. Biol. 2014, 22, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Zhu, L.; Schwaneberg, U. Directed evolution 2.0: Improving and deciphering enzyme properties. Chem. Commun. 2015, 51, 9760–9772. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Computer-aided protein directed evolution: A review of web servers, databases and other computational tools for protein engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209008. [Google Scholar] [CrossRef] [Green Version]
- Lutz, S. Beyond directed evolution—Semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2010, 21, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Romero-Rivera, A.; Garcia-Borras, M.; Osuna, S. Computational tools for the evaluation of laboratory-engineered biocatalysts. Chem. Commun. 2016, 53, 284–297. [Google Scholar] [CrossRef] [Green Version]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Chica, R.A.; Doucet, N.; Pelletier, J.N. Semi-rational approaches to engineering enzyme activity: Combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005, 16, 378–384. [Google Scholar] [CrossRef]
- Lee, H.-L.; Chang, C.-K.; Jeng, W.-Y.; Wang, A.H.-J.; Liang, P.-H. Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Eng. Des. Sel. 2012, 25, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, W.; Ji, P.; Zhao, Y.; Hua, C.; Han, C. Engineering the conserved and noncatalytic residues of a thermostable β-1, 4-endoglucanase to improve specific activity and thermostability. Sci. Rep. 2018, 8, 2954. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Tu, T.; Wang, X.; Wang, Y.; Ma, R.; Su, X.; Xie, X.; Yao, B.; Luo, H. Enhancing the catalytic activity of a novel GH5 cellulase Gt Cel5 from Gloeophyllum trabeum CBS 900.73 by site-directed mutagenesis on loop 6. Biotechnol. Biofuels 2018, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, J.M.; Azócar, M.A.; Rodríguez, V.; Ramírez-Sarmiento, C.A.; Andrews, B.A.; Asenjo, J.A.; Parra, L.P. Relevance of Local Flexibility Near the Active Site for Enzymatic Catalysis: Biochemical Characterization and Engineering of Cellulase Cel5A From Bacillus agaradherans. Biotechnol. J. 2018, 13, 1700669. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Goswami, S.; Das, S.; Datta, S. Exploiting non-conserved residues to improve activity and stability of Halothermothrix orenii β-glucosidase. Appl. Microbiol. Biotechnol. 2017, 101, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Li, W.; Hua, C.; Sun, F.; Bi, P.; Wang, Q. Enhancement of catalytic activity and thermostability of a thermostable cellobiohydrolase from Chaetomium thermophilum by site-directed mutagenesis. Int. J. Biol. Macromol. 2018, 116, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Sumitani, J.-I.; Tanaka, K.; Tani, S.; Kawaguchi, T. Site-saturation mutagenesis for β-glucosidase 1 from Aspergillus aculeatus to accelerate the saccharification of alkaline-pretreated bagasse. Appl. Microbiol. Biotechnol. 2016, 100, 10495–10507. [Google Scholar] [CrossRef]
- Taylor, L.E.; Knott, B.C.; Baker, J.O.; Alahuhta, P.M.; Hobdey, S.E.; Linger, J.G.; Lunin, V.V.; Amore, A.; Subramanian, V.; Podkaminer, K. Engineering enhanced cellobiohydrolase activity. Nat. Commun. 2018, 9, 1186. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Gu, J.; Xie, W.; Yu, H. Directed co-evolution of an endoglucanase and a β-glucosidase in Escherichia coli by a novel high-throughput screening method. Chem. Commun. 2013, 49, 7219–7221. [Google Scholar] [CrossRef]
- Larue, K.; Melgar, M.; Martin, V.J. Directed evolution of a fungal β-glucosidase in Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Hardiman, E.; Gibbs, M.; Reeves, R.; Bergquist, P. Directed evolution of a thermophilic β-glucosidase for cellulosic bioethanol production. Appl. Biochem. Biotechnol. 2010, 161, 301–312. [Google Scholar] [CrossRef]
- Liang, C.; Fioroni, M.; Rodríguez-Ropero, F.; Xue, Y.; Schwaneberg, U.; Ma, Y. Directed evolution of a thermophilic endoglucanase (Cel5A) into highly active Cel5A variants with an expanded temperature profile. J. Biotechnol. 2011, 154, 46–53. [Google Scholar] [CrossRef]
- Lin, L.; Fu, C.; Huang, W. Improving the activity of the endoglucanase, Cel8M from Escherichia coli by error-prone PCR. Enzym. Microb. Technol. 2016, 86, 52–58. [Google Scholar] [CrossRef]
- Liu, M.; Xie, W.; Xu, H.; Gu, J.; Lv, X.; Yu, H.; Ye, L. Directed evolution of an exoglucanase facilitated by a co-expressed β-glucosidase and construction of a whole engineered cellulase system in Escherichia coli. Biotechnol. Lett. 2014, 36, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- Longwell, C.K.; Labanieh, L.; Cochran, J.R. High-throughput screening technologies for enzyme engineering. Curr. Opin. Biotechnol. 2017, 48, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Körfer, G.; Pitzler, C.; Vojcic, L.; Martinez, R.; Schwaneberg, U. In vitro flow cytometry-based screening platform for cellulase engineering. Sci. Rep. 2016, 6, 26128. [Google Scholar] [CrossRef] [Green Version]
- Schiano-di-Cola, C.; Røjel, N.; Jensen, K.; Kari, J.; Sørensen, T.H.; Borch, K.; Westh, P. Systematic deletions in the cellobiohydrolase (CBH) Cel7A from the fungus Trichoderma reesei reveal flexible loops critical for CBH activity. J. Biol. Chem. 2019, 294, 1807–1815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.A.; Morais, M.A.; Terrett, O.M.; Lyczakowski, J.J.; Zanphorlin, L.M.; Ferreira-Filho, J.A.; Tonoli, C.C.; Murakami, M.T.; Dupree, P.; Souza, A.P. An engineered GH1 β-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials. Sci. Rep. 2019, 9, 4903. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xue, Y.; Lin, Y. Enhanced catalytic efficiency in quercetin-4′-glucoside hydrolysis of Thermotoga maritima β-glucosidase A by site-directed mutagenesis. J. Agric. Food Chem. 2014, 62, 6763–6770. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Joo, J.E.; Jeon, S.D.; Hyeon, J.E.; Kim, S.W.; Um, Y.S.; Han, S.O. Enhanced thermostability of mesophilic endoglucanase Z with a high catalytic activity at active temperatures. Int. J. Biol. Macromol. 2016, 86, 269–276. [Google Scholar] [CrossRef]
- Ito, Y.; Ikeuchi, A.; Imamura, C. Advanced evolutionary molecular engineering to produce thermostable cellulase by using a small but efficient library. Protein Eng. Des. Sel. 2012, 26, 73–79. [Google Scholar] [CrossRef]
- Wu, I.; Heel, T.; Arnold, F.H. Role of cysteine residues in thermal inactivation of fungal Cel6A cellobiohydrolases. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1539–1544. [Google Scholar] [CrossRef] [PubMed]
- Anbar, M.; Gul, O.; Lamed, R.; Sezerman, U.O.; Bayer, E.A. Improved thermostability of Clostridium thermocellum endoglucanase Cel8A by using consensus-guided mutagenesis. Appl. Environ. Microbiol. 2012, 78, 3458–3464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-J.; Hsiao, Y.-Y.; Chen, Y.-P.; Ma, T.-Y.; Tseng, C.-P. Polarity Alteration of a Calcium Site Induces a Hydrophobic Interaction Network and Enhances Cel9A Endoglucanase Thermostability. Appl. Environ. Microbiol. 2016, 82, 1662–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayram Akcapinar, G.; Venturini, A.; Martelli, P.L.; Casadio, R.; Sezerman, U.O. Modulating the thermostability of Endoglucanase I from Trichoderma reesei using computational approaches. Protein Eng. Des. Sel. 2015, 28, 127–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Shi, H.; Xu, L.; Zhu, X.; Li, X. Site-directed mutagenesis of a hyperthermophilic endoglucanase Cel12B from Thermotoga maritima based on rational design. PLoS ONE 2015, 10, e0133824. [Google Scholar]
- Chokhawala, H.A.; Roche, C.M.; Kim, T.-W.; Atreya, M.E.; Vegesna, N.; Dana, C.M.; Blanch, H.W.; Clark, D.S. Mutagenesis of Trichoderma reesei endoglucanase I: Impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol. 2015, 15, 11. [Google Scholar] [CrossRef] [Green Version]
- Badieyan, S.; Bevan, D.R.; Zhang, C. Study and design of stability in GH5 cellulases. Biotechnol. Bioeng. 2012, 109, 31–44. [Google Scholar] [CrossRef]
- Telke, A.A.; Zhuang, N.; Ghatge, S.S.; Lee, S.-H.; Shah, A.A.; Khan, H.; Um, Y.; Shin, H.-D.; Chung, Y.R.; Lee, K.H. Engineering of family-5 glycoside hydrolase (Cel5A) from an uncultured bacterium for efficient hydrolysis of cellulosic substrates. PLoS ONE 2013, 8, e65727. [Google Scholar] [CrossRef]
- Voutilainen, S.P.; Nurmi-Rantala, S.; Penttilä, M.; Koivula, A. Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014, 98, 2991–3001. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Wu, Z.-F.; Chen, H.; Lai, X.; Han, X.-Y.; Wu, Q. Characterization of novel EGs reconstructed from Bacillus subtilis endoglucanase. Appl. Biochem. Biotechnol. 2013, 169, 1764–1773. [Google Scholar]
- Moraïs, S.; Stern, J.; Kahn, A.; Galanopoulou, A.P.; Yoav, S.; Shamshoum, M.; Smith, M.A.; Hatzinikolaou, D.G.; Arnold, F.H.; Bayer, E.A. Enhancement of cellulosome-mediated deconstruction of cellulose by improving enzyme thermostability. Biotechnol. Biofuels 2016, 9, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.A.; Rentmeister, A.; Snow, C.D.; Wu, T.; Farrow, M.F.; Mingardon, F.; Arnold, F.H. A diverse set of family 48 bacterial glycoside hydrolase cellulases created by structure-guided recombination. FEBS J. 2012, 279, 4453–4465. [Google Scholar] [CrossRef] [PubMed]
- Heinzelman, P.; Snow, C.D.; Wu, I.; Nguyen, C.; Villalobos, A.; Govindarajan, S.; Minshull, J.; Arnold, F.H. A family of thermostable fungal cellulases created by structure-guided recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 5610–5615. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhang, X.-Z.; Zhang, Z.; Zhang, Y.-H.P. Engineering of Clostridium phytofermentans endoglucanase Cel5A for improved thermostability. Appl. Environ. Microbiol. 2010, 76, 4914–4917. [Google Scholar] [CrossRef] [Green Version]
- Anbar, M.; Lamed, R.; Bayer, E.A. Thermostability enhancement of Clostridium thermocellum cellulosomal endoglucanase Cel8A by a single glycine substitution. ChemCatChem 2010, 2, 997–1003. [Google Scholar] [CrossRef]
- Dana, C.M.; Saija, P.; Kal, S.M.; Bryan, M.B.; Blanch, H.W.; Clark, D.S. Biased clique shuffling reveals stabilizing mutations in cellulase Cel7A. Biotechnol. Bioeng. 2012, 109, 2710–2719. [Google Scholar] [CrossRef]
- Chang, C.-J.; Lee, C.-C.; Chan, Y.-T.; Trudeau, D.L.; Wu, M.-H.; Tsai, C.-H.; Yu, S.-M.; Ho, T.-H.D.; Wang, A.H.-J.; Hsiao, C.-D.; et al. Exploring the mechanism responsible for cellulase thermostability by structure-guided recombination. PLoS ONE 2016, 11, e0147485. [Google Scholar] [CrossRef]
- Goedegebuur, F.; Dankmeyer, L.; Gualfetti, P.; Karkehabadi, S.; Hansson, H.; Jana, S.; Huynh, V.; Kelemen, B.R.; Kruithof, P.; Larenas, E.A.; et al. Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution. J. Biol. Chem. 2017, 292, 17418–17430. [Google Scholar] [CrossRef] [Green Version]
- Wijma, H.J.; Floor, R.J.; Jekel, P.A.; Baker, D.; Marrink, S.J.; Janssen, D.B. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 2014, 27, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Khersonsky, O.; Lipsh, R.; Avizemer, Z.; Ashani, Y.; Goldsmith, M.; Leader, H.; Dym, O.; Rogotner, S.; Trudeau, D.L.; Prilusky, J. Automated design of efficient and functionally diverse enzyme repertoires. Mol. Cell 2018, 72, 178–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathi, P.C.; Mulnaes, D.; Gohlke, H. VisualCNA: A GUI for interactive constraint network analysis and protein engineering for improving thermostability. Bioinformatics 2015, 31, 2394–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoav, S.; Stern, J.; Salama-Alber, O.; Frolow, F.; Anbar, M.; Karpol, A.; Hadar, Y.; Morag, E.; Bayer, E.A. Directed Evolution of Clostridium thermocellum β-Glucosidase A Towards Enhanced Thermostability. Int. J. Mol. Sci. 2019, 20, 4701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashirova, A.; Pramanik, S.; Volkov, P.; Rozhkova, A.; Nemashkalov, V.; Zorov, I.; Gusakov, A.; Sinitsyn, A.; Schwaneberg, U.; Davari, M.D. Disulfide bond engineering of an endoglucanase from Penicillium verruculosum to improve its thermostability. Int. J. Mol. Sci. 2019, 20, 1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, F.; Vermaas, J.V.; Zheng, J.; Wang, Y.; Tu, T.; Wang, X.; Xie, X.; Yao, B.; Beckham, G.T.; Luo, H. Activity and Thermostability of GH5 Endoglucanase Chimeras from Mesophilic and Thermophilic Parents. Appl. Environ. Microbiol. 2019, 85, e02079-18. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Li, S.; Huang, X.; Qin, Z.; Kong, W.; Xie, W.; Liu, Y. Enhancing the thermostability of highly active and glucose-tolerant β-Glucosidase Ks5A7 by directed evolution for good performance of three properties. J. Agric. Food Chem. 2018, 66, 13228–13235. [Google Scholar] [CrossRef]
- Zhou, C.; Xue, Y.; Ma, Y. Evaluation and directed evolution for thermostability improvement of a GH 13 thermostable α-glucosidase from Thermus thermophilus TC11. BMC Biotechnol. 2015, 15, 97. [Google Scholar] [CrossRef] [Green Version]
- Mitrovic, A.; Flicker, K.; Steinkellner, G.; Gruber, K.; Reisinger, C.; Schirrmacher, G.; Camattari, A.; Glieder, A. Thermostability improvement of endoglucanase Cel7B from Hypocrea pseudokoningii. J. Mol. Catal. B Enzym. 2014, 103, 16–23. [Google Scholar] [CrossRef]
- Wu, I.; Arnold, F.H. Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnol. Bioeng. 2013, 110, 1874–1883. [Google Scholar] [CrossRef]
- Smith, M.A.; Bedbrook, C.N.; Wu, T.; Arnold, F.H. Hypocrea jecorina cellobiohydrolase I stabilizing mutations identified using noncontiguous recombination. ACS Synth. Biol. 2013, 2, 690–696. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-J.; Peng, Y.-J.; Zhang, L.-Q.; Li, A.-N.; Li, D.-C. Directed evolution and structural prediction of cellobiohydrolase II from the thermophilic fungus Chaetomium thermophilum. Appl. Microbiol. Biotechnol. 2012, 95, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Heinzelman, P.; Komor, R.; Kanaan, A.; Romero, P.; Yu, X.; Mohler, S.; Snow, C.; Arnold, F. Efficient screening of fungal cellobiohydrolase class I enzymes for thermostabilizing sequence blocks by SCHEMA structure-guided recombination. Protein Eng. Des. Sel. 2010, 23, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Voutilainen, S.P.; Murray, P.G.; Tuohy, M.G.; Koivula, A. Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng. Des. Sel. 2009, 23, 69–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, G.; Lewis, J.C. Introduction: Biocatalysis in industry. Chem. Rev. 2018, 118, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazlauskas, R. Engineering more stable proteins. Chem. Soc. Rev. 2018, 47, 9026–9045. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys. Condens. Matter 2010, 22, 323101. [Google Scholar] [CrossRef]
- Britton, J.; Majumdar, S.; Weiss, G.A. Continuous flow biocatalysis. Chem. Soc. Rev. 2018, 47, 5891–5918. [Google Scholar] [CrossRef]
- Swatloski, R.P.; Spear, S.K.; Holbrey, J.D.; Rogers, R.D. Dissolution of cellose with ionic liquids. J. Am. Chem. Soc. 2002, 124, 4974–4975. [Google Scholar] [CrossRef]
- Socha, A.M.; Parthasarathi, R.; Shi, J.; Pattathil, S.; Whyte, D.; Bergeron, M.; George, A.; Tran, K.; Stavila, V.; Venkatachalam, S. Efficient biomass pretreatment using ionic liquids derived from lignin and hemicellulose. Proc. Natl. Acad. Sci. USA 2014, 111, E3587–E3595. [Google Scholar] [CrossRef] [Green Version]
- Guerriero, G.; Hausman, J.F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Lignocellulosic biomass: Biosynthesis, degradation, and industrial utilization. Eng. Life Sci. 2016, 16, 1–16. [Google Scholar] [CrossRef]
- Jaeger, V.; Burney, P.; Pfaendtner, J. Comparison of three ionic liquid-tolerant cellulases by molecular dynamics. Biophys. J. 2015, 108, 880–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordwald, E.M.; Brunecky, R.; Himmel, M.E.; Beckham, G.T.; Kaar, J.L. Charge engineering of cellulases improves ionic liquid tolerance and reduces lignin inhibition. Biotechnol. Bioeng. 2014, 111, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Wolski, P.W.; Dana, C.M.; Clark, D.S.; Blanch, H.W. Engineering ionic liquid-tolerant cellulases for biofuels production. Protein Eng. Des. Sel. 2016, 29, 117–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pottkämper, J.; Barthen, P.; Ilmberger, N.; Schwaneberg, U.; Schenk, A.; Schulte, M.; Ignatiev, N.; Streit, W.R. Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green Chem. 2009, 11, 957–965. [Google Scholar] [CrossRef]
- Lehmann, C.; Sibilla, F.; Maugeri, Z.; Streit, W.R.; de María, P.D.; Martinez, R.; Schwaneberg, U. Reengineering CelA2 cellulase for hydrolysis in aqueous solutions of deep eutectic solvents and concentrated seawater. Green Chem. 2012, 14, 2719–2726. [Google Scholar] [CrossRef]
- Lehmann, C.; Bocola, M.; Streit, W.R.; Martinez, R.; Schwaneberg, U. Ionic liquid and deep eutectic solvent-activated CelA2 variants generated by directed evolution. Appl. Microbiol. Biotechnol. 2014, 98, 5775–5785. [Google Scholar] [CrossRef]
- Chen, Z.; Pereira, J.H.; Liu, H.; Tran, H.M.; Hsu, N.S.; Dibble, D.; Singh, S.; Adams, P.D.; Sapra, R.; Hadi, M.Z. Improved activity of a thermophilic cellulase, Cel5A, from Thermotoga maritima on ionic liquid pretreated switchgrass. PLoS ONE 2013, 8, e79725. [Google Scholar] [CrossRef] [Green Version]
- Warden, A.C.; Williams, M.; Peat, T.S.; Seabrook, S.A.; Newman, J.; Dojchinov, G.; Haritos, V.S. Rational engineering of a mesohalophilic carbonic anhydrase to an extreme halotolerant biocatalyst. Nat. Commun. 2015, 6, 10278. [Google Scholar] [CrossRef]
- Hess, B.; van der Vegt, N.F. Cation specific binding with protein surface charges. Proc. Natl. Acad. Sci. USA 2009, 106, 13296–13300. [Google Scholar] [CrossRef] [Green Version]
- Yamamura, A.; Ichimura, T.; Kamekura, M.; Mizuki, T.; Usami, R.; Makino, T.; Ohtsuka, J.; Miyazono, K.-I.; Okai, M.; Nagata, K. Molecular mechanism of distinct salt-dependent enzyme activity of two halophilic nucleoside diphosphate kinases. Biophys. J. 2009, 96, 4692–4700. [Google Scholar] [CrossRef] [Green Version]
- Park, C.; Raines, R.T. Quantitative analysis of the effect of salt concentration on enzymatic catalysis. J. Am. Chem. Soc. 2001, 123, 11472–11479. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Dong, Y.; Hong, X.; Pang, X.; Chen, D.; Chen, X. Directed Evolution of Dunaliella salina Ds-26-16 and Salt-Tolerant Response in Escherichia coli. Int. J. Mol. Sci. 2016, 17, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gribenko, A.V.; Patel, M.M.; Liu, J.; McCallum, S.A.; Wang, C.; Makhatadze, G.I. Rational stabilization of enzymes by computational redesign of surface charge–charge interactions. Proc. Natl. Acad. Sci. USA 2009, 106, 2601–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Datta, S.; Eichler, J.; Ivanova, N.; Axen, S.D.; Kerfeld, C.A.; Chen, F.; Kyrpides, N.; Hugenholtz, P.; Cheng, J.-F. Identification of a haloalkaliphilic and thermostable cellulase with improved ionic liquid tolerance. Green Chem. 2011, 13, 2083–2090. [Google Scholar] [CrossRef] [Green Version]
- Gaur, R.; Tiwari, S. Isolation, production, purification and characterization of an organic-solvent-thermostable alkalophilic cellulase from Bacillus vallismortis RG-07. BMC Biotechnol. 2015, 15, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barruetabeña, N.; Alonso-Lerma, B.; Galera-Prat, A.; Joudeh, N.; Barandiaran, L.; Aldazabal, L.; Arbulu, M.; Alcalde, M.; De Sancho, D.; Gavira, J.A. Resurrection of efficient Precambrian endoglucanases for lignocellulosic biomass hydrolysis. Commun. Chem. 2019, 2, 76. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, H.; Okada, K.; Onodera, T.; Ogasawara, W.; Okada, H.; Morikawa, Y. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl. Microbiol. Biotechnol. 2009, 83, 649–657. [Google Scholar] [CrossRef]
- Qin, Y.; Wei, X.; Song, X.; Qu, Y. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. J. Biotechnol. 2008, 135, 190–195. [Google Scholar] [CrossRef]
- Lin, H.; Li, W.; Guo, C.; Qu, S.; Ren, N. Advances in the study of directed evolution for cellulases. Front. Environ. Sci. Eng. China 2011, 5, 519–525. [Google Scholar] [CrossRef]
- Xia, W.; Xu, X.; Qian, L.; Shi, P.; Bai, Y.; Luo, H.; Ma, R.; Yao, B. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol. Biofuels 2016, 9, 147. [Google Scholar] [CrossRef] [Green Version]
- Dana, C.M.; Dotson-Fagerstrom, A.; Roche, C.M.; Kal, S.M.; Chokhawala, H.A.; Blanch, H.W.; Clark, D.S. The importance of pyroglutamate in cellulase Cel7A. Biotechnol. Bioeng. 2014, 111, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Buß, O.; Rudat, J.; Ochsenreither, K. FoldX as protein engineering tool: Better than random based approaches? Comput. Struct. Biotechnol. J. 2018, 16, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Harman, G.E. Trichoderma and Gliocladium. Volume 1: Basic Biology, Taxonomy and Genetics; Taylor and Francis Ltd.: Abingdon, UK, 1998. [Google Scholar]
- Chemudupati, M.; Johns, M.; Osmani, S.A. The mode of mitosis is dramatically modified by deletion of a single nuclear pore complex gene in Aspergillus nidulans. Fungal Genet. Biol. 2019, 130, 72–81. [Google Scholar] [CrossRef]
- Szakács, G.; Réczey, K.; Hernádi, P.; Dobozi, M. Penicillium verruculosum WA 30 a new source of cellulase. Eur. J. Appl. Microbiol. Biotechnol. 1981, 11, 120–124. [Google Scholar] [CrossRef]
- Sinitsyn, A.P.; Rozhkova, A.M. Penicillium canescens host as the platform for development of a new recombinant strain producers of carbohydrases. In Microorganisms in Biorefineries; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–19. [Google Scholar]




| Cellulase (Source) | Improvement | Engineering Method | Activity Assay | Molecular Effect | Reference |
|---|---|---|---|---|---|
| Cellobiohydrolase TreCel7A (Trichoderma reseii) | N.A. | Rational design—Consensus mutations, loop engineering | 1pNPL 2MCC/ 3PAHBAH | B2 loop is important in the activity of Cel7A | [94] |
| β-glucosidase GH1 (Trichoderma harzianum) | 5-fold improvement in catalytic efficiency and 3-fold activity improvement in the presence of 0.2 M glucose | Rational design—selection of gate keeper amino acids | 4pNPG | Narrowing of active-site pocket | [95] |
| Endoglucanase Cel5A (Bacillus agaradherans) | 3-fold improvement of activity at 5 °C | Rational design— MSA and MD, loop engineering | 5pNPC | Decrease in Ea, ΔH#, and ΔS# is concomitant with a higher flexibility of the active site region | [81] |
| Cellobiohydrolase PfCel7A (Penicillium funiculosum) | Improvement of >60% in terms of the time to 80% conversion of PASC | Rational design—Chimera library with homologous CBH I | 1pNPL and Biomass/ 6HPLC | Increased ligand and structural flexibility at the binding tunnel entrance | [85] |
| Endoglucanase GtCel5 (Gloeophyllum trabeum) | Increase in specific activity of 1.7-fold toward barley β-glucan | Rational design— MSA of loops | 2CMC/ 7DNS | Effects on the local hydrogen-bonding network produce stronger interaction with the substrate | [80] |
| Cellobiohydrolase CtCel6 (Chaetomium thermophilum) | Increased by 1.82-, 1.65-, and 1.43-fold against β-D-glucan, PASC and CMC-Na, respectively | Rational design—Homology analysis | β-glucan, 2CMC, 8PASC/ 7DNS | N.A. | [83] |
| Endoglucanase CTendo45 (Chaetomium thermophilum) | 4-fold increase in kcat and 1.94-fold in catalytic efficiency | Rational design—Conserved and non-catalytic residues | β-glucan, 2CMC/ 7DNS | Decrease in entropy and disruption in of internal electrostatic interactions | [79] |
| β-glucosidase (Halothermothrix orenii) | 1.8-fold increase in turnover number with the natural substrate | Rational design— MSA. Selection of non-conserved residues near entrance loop | 4pNPG and cellobiose/ 9GOD-POD assay kit | Reduction of water contact and steric impairment | [82] |
| Endoglucanase Cel8M (Escherichia coli) | 1.6-fold increased specific activity | Directed evolution— ep-PCR | Congo red-2CMC/ 7DNS | Formation of a hydrogen network involved in the substrate binding | [90] |
| CelA2 (Metagenomic library GenBank: JF826524.1) | 13.3-fold improvement in specific activity | Directed evolution— ep-PCR | 10FDC/ fluorescein and 114-MUC | N.A. | [93] |
| β-glucosidase 1 AaBGL1 (Aspergillus aculeatus) | 2.7 times higher kcat/Km toward cellobiose | Semi-rational design—SSM in amino acids of subsite +1 | cellobiose/ Glucose CII-Test Wako Alkaline-pre-treated bagasse/ 12HPAEC-PAD | N.A. | [84] |
| β-glucosidase BGL1 (Aspergillus niger) | 3.3-fold improvement in the Vmax with the natural substrate | Directed evolution— ep-PCR | 4pNPG and cellobiose/ 9GOD-POD assay kit | N.A. | [87] |
| Cellobiohydrolase CBH A (Cellulomonas fimi) | 2.7-fold improvement of the specific activity | Directed evolution— ep-PCR | cellulose/ β-glucosidase | N.A. | [91] |
| β-glucosidase A (Thermothoga maritima) | 1.6-fold improvement of kcat/Km toward pNPG | Rational design—Conserved amino acids of subsite −1 and docking | 4pNPG | N.A. | [96] |
| Endoglucanase CenA (Cellulomonas fimi) | 2.7-fold increase in specific activity | Directed evolution—ep-PCR | Whatman no. 1 filter paper with a coupled with 9GOD-POD | N.A. | [86] |
| β-glucosidase TrBgl2 (Trichoderma reseii) | 4.6-fold increase in kcat and 5.3-fold improve in kcat/Km (277 U/mg) | Rational design—MSA of substrate entrance cleft | 4pNPG | better interaction of the substrate with the active site | [78] |
| Endoglucanase Cel5A (Thermoanaerobacter tengcongensis) | 1.9- and 1.78-fold improvement in specific activity and catalytic efficiency, respectively | Directed evolution—ep-PCR | Congo red, 2CMC/ 7DNS | Loss of hydrogen bond network | [89] |
| β-glucosidase A bglA (Caldicellulosiruptor saccharolyticus) | 1.8- and 1.7-fold improvement in specific activity towards artificial and natural substrate, respectively | Directed evolution—ep-PCR | 4pNPG, 1pNPL and cellobiose/ Ample red coupled with 9GOD-POD Assay Kit | N.A. | [88] |
| Cellulase (Source) | Non-Conventional Media | Improvement | Engineering Method | Activity Assay | Molecular Effect | Reference |
|---|---|---|---|---|---|---|
| Cellobiohydrolase Cel7A (Talaromyces emersonii) | [MMIM][DMP] and [EMIM][Ac] | 3-fold | Semi-rational design—DNA shuffling biased clique shuffling | ILs-treated 1MCC assay | N.A. | [140] |
| CelA2 (metagenome, GenBank: JF826524.1) | 3-fold concentrated seawater | 1.6-fold | Directed evolution—ep-PCR with PLICing | 24-MUC | N.A. | [142] |
| CelA2 (metagenome, GenBank: KC964209) | [BMIM][Cl] | 23-fold | Directed evolution—ep-PCR | 24-MUC | Salt bridge formation D287 and R300 in variant (H288F- S300R) | [143] |
| Cellulase cocktail (Trichoderma reesei) | [BMIM][Cl] | 2-fold | Rational design—Succinyl induced charge modification | ILs-treated 1MCC | Succinylation preferential lead to the exclusion of the Cl− | [139] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras, F.; Pramanik, S.; M. Rozhkova, A.; N. Zorov, I.; Korotkova, O.; P. Sinitsyn, A.; Schwaneberg, U.; D. Davari, M. Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails. Int. J. Mol. Sci. 2020, 21, 1589. https://doi.org/10.3390/ijms21051589
Contreras F, Pramanik S, M. Rozhkova A, N. Zorov I, Korotkova O, P. Sinitsyn A, Schwaneberg U, D. Davari M. Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails. International Journal of Molecular Sciences. 2020; 21(5):1589. https://doi.org/10.3390/ijms21051589
Chicago/Turabian StyleContreras, Francisca, Subrata Pramanik, Aleksandra M. Rozhkova, Ivan N. Zorov, Olga Korotkova, Arkady P. Sinitsyn, Ulrich Schwaneberg, and Mehdi D. Davari. 2020. "Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails" International Journal of Molecular Sciences 21, no. 5: 1589. https://doi.org/10.3390/ijms21051589