Autoregulation of greA Expression Relies on GraL Rather than on greA Promoter Region
Abstract
:1. Introduction
2. Results
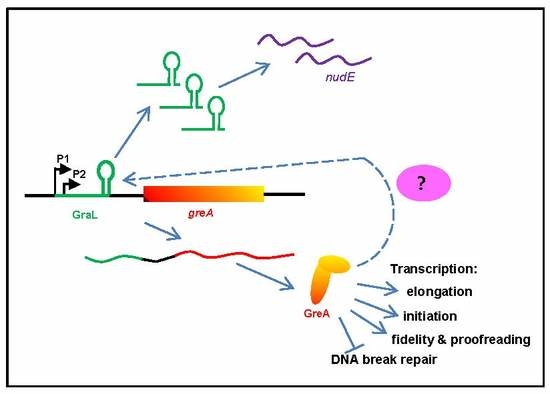
2.1. GreA Autoregulation Depends on Catalytically Active GreA
2.2. GreA Autoregulation is Independent of the Promoter Region
2.3. The Amount of GraL Produced Depends on the GreA Level in the Cell
2.4. The Observed GreA Inhibition of greA Expression is not Due to GraL Acting in trans on its Own mRNA
2.5. GreA Mediated Regulation Requires Cis-Acting Gral Sequences in Addition to the Terminator Structure
2.6. None of the Nus Factors nor σE Affect greA Regulation
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. β-Galactosidase Assays
4.3. Northern Blots
4.4. Single Round in vitro Transcription
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IPTG | Isopropyl-β-D-thio-galactopyranoside |
| RNAP | RNA polymerase |
| sRNA | small RNA |
| UTR | untranslated region |
| nt | nucleotides |
| wt | wild type |
References
- Sen, R.; Chalissery, J.; Muteeb, G. Nus Factors of Escherichia coli. EcoSal Plus 2014, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Merino, E.; Yanofsky, C. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends Genet. 2005, 21, 260–264. [Google Scholar] [CrossRef] [PubMed]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef] [PubMed]
- Sedlyarova, N.; Rescheneder, P.; Magán, A.; Popitsch, N.; Rziha, N.; Bilusic, I.; Epshtein, V.; Zimmermann, B.; Lybecker, M.; Sedlyarov, V.; et al. Natural RNA polymerase aptamers regulate transcription in E. coli. Mol. Cell 2017, 67, 30–43. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3, a003798. [Google Scholar] [CrossRef]
- Carrier, M.C.; Lalaouna, D.; Massé, E. Broadening the definition of bacterial small RNAs: Characteristics and mechanisms of action. Annu. Rev. Microbiol. 2018, 72, 141–161. [Google Scholar] [CrossRef]
- Komissarova, N.; Kashlev, M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. USA 1997, 94, 1755–1780. [Google Scholar] [CrossRef]
- Mustaev, A.; Roberts, J.; Gottesman, M. Transcription elongation. Transcription 2017, 8, 150–161. [Google Scholar] [CrossRef] [Green Version]
- Borukhov, S.; Lee, J.; Laptenko, O. Bacterial transcription elongation factors: New insights into molecular mechanism of action. Mol. Microbiol. 2005, 55, 1315–1324. [Google Scholar] [CrossRef]
- Zhang, G.; Campbell, E.A.; Minakhin, L.; Richter, C.; Severinov, K.; Darst, S.A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 1999, 98, 811–824. [Google Scholar] [CrossRef]
- Vassylyev, D.G.; Sekine, S.; Laptenko, O.; Lee, J.; Vassylyeva, M.N.; Borukhov, S.; Yokoyama, S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 2002, 417, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Lee, J.; Lomakin, I.; Borukhov, S. Transcript cleavage factors GreA and GreB act as a transient catalytic components of RNA polymerase. EMBO J. 2003, 22, 6322–6334. [Google Scholar] [CrossRef] [PubMed]
- Nickels, B.E.; Hochschild, A. Regulation of RNA polymerase through the secondary channel. Cell 2004, 118, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Blankschein, M.D.; Potrykus, K.; Grace, E.; Choudhary, A.; Vinella, D.; Cashel, M.; Herman, C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2008, 5, e1000345. [Google Scholar] [CrossRef] [PubMed]
- Lamour, V.; Hogan, B.P.; Erie, D.A.; Darst, S.A. Crystalstructure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J. Mol. Biol. 2006, 356, 179–188. [Google Scholar] [CrossRef]
- Potrykus, K.; Vinella, D.; Murphy, H.; Szalewska-Palasz, A.; D’Ari, R.; Cashel, M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006, 281, 15238–15248. [Google Scholar] [CrossRef]
- Aberg, A.; Shingler, V.; Balsalobre, C. Regulation of the fimB promoter: A case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 2008, 67, 1223–1241. [Google Scholar] [CrossRef]
- Vinella, D.; Potrykus, K.; Murphy, H.; Cashel, M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bact. 2012, 194, 261–273. [Google Scholar] [CrossRef]
- Fernandez-Coll, L.; Potrykus, K.; Cashel, M. Puzzling conformational changes affecting proteins binding to the RNA polymerase. Proc. Natl. Acad. Sci. USA 2018, 115, 12550–12552. [Google Scholar] [CrossRef] [Green Version]
- Hsu, L.M.; Vo, N.V.; Chamberlin, M.J. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl. Acad. Sci. USA 1995, 92, 11588–11592. [Google Scholar] [CrossRef]
- Bubunenko, M.G.; Rattray, A.J.; Gotte, D.R.; Kireeva, M.L.; Irizarry-Caro, J.A.; Li, X.; Jin, D.J.; Court, D.L.; Strathern, J.N.; Kashlev, M. A Cre transcription fidelity reporter identifies GreA as a major RNA proofreading factor in Escherichia coli. Genetics 2017, 206, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Traverse, C.C.; Ochman, H. A genome-wide assay specifies only GreA as a transcription fidelity factor in Escherichia coli. G3: Genes Genomes Genet. 2018, 8, 2257–2264. [Google Scholar] [CrossRef] [PubMed]
- Gaviria-Cantin, T.; El Mouali, Y.; Le Guyon, S.; Römling, U.; Balsalobre, C. Gre factors-mediated control of hilD transcription is essential for the invasion of epithelial cells by Salmonella enterica serovar Typhimurium. PLoS Pathog. 2017, 13, e1006312. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Peterson, S.N.; Gill, S.R.; Cline, R.T.; White, O.; Fraser, C.M.; Smith, H.O.; Venter, J.C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 1999, 286, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, K.; Murphy, H.; Chen, J.; Epstein, J.; Cashel, M. Imprecise transcription termination within Escherichia coli greA leader gives rise to an array of short transcripts, GraL. Nucleic Acids Res. 2010, 38, 1636–1651. [Google Scholar] [CrossRef] [PubMed]
- Dylewski, M.; Ćwiklińska, M.; Potrykus, K. A search for the in trans role of GraL, an Escherichia coli small RNA. Acta Biochim. Pol. 2018, 65, 141–149. [Google Scholar] [CrossRef]
- Hirschel, B.J.; Shen, V.; Schlessinger, D. Lactose operon transcription from wild-type and L8-UV5 lac promoters in Escherichia coli treated with chloramphenicol. J. Bact. 1980, 143, 1534–1537. [Google Scholar]
- Peters, J.M.; Vangeloff, A.D.; Landick, R. Bacterial transcription terminators: The RNA 3′-end chronicles. J. Mol. Biol. 2011, 412, 793–813. [Google Scholar] [CrossRef]
- Chen, J.; Morita, T.; Gottesman, S. Regulation of transcription termination of small RNAs and by small RNAs: Molecular mechanisms and biological functions. Front. Cell. Infect. Microbiol. 2019, 9, 201. [Google Scholar] [CrossRef]
- Walsh, N.P.; Alba, B.M.; Bose, B.; Gross, C.A.; Sauer, R.T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003, 113, 61–71. [Google Scholar] [CrossRef]
- Sivaramakrishnan, P.; Sepúlveda, L.A.; Halliday, J.A.; Liu, J.; Núñez, M.A.B.; Golding, I.; Rosenberg, S.M.; Herman, C. The transcription fidelity factor GreA impedes DNA break repair. Nature 2017, 550, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.; Lee, J.; Ozerova, M.; Semenova, E.; Datsenko, K.; Wanner, B.L.; Severinov, K.; Borukhov, S. Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bact. 2007, 189, 8772–8785. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Lemke, J.J.; Vrentas, C.E.; Gaal, T.; Ross, W.; Gourse, R.L. Effects of DksA, GreA, and GreB on transcription initiation: Insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 2007, 366, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Loh, E.; Dussurget, O.; Gripenland, J.; Vaitkevicius, K.; Tiensuu, T.; Mandin, P.; Repoila, F.; Buchriesier, C.; Cossart, P.; Johansson, J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 2009, 139, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Tu, N.; Carroll, R.K.; Weiss, A.; Shaw, L.N.; Nicolas, G.; Thomas, S.; Lima, A.; Okaro, U.; Anderson, B. A family of genus-specific RNAs in tandem with DNA-binding proteins control expression of the badA major virulence factor gene in Bartonella henselae. Microbiol. Open 2017, 6, e00420. [Google Scholar] [CrossRef]
- Naville, M.; Gautheret, D. Premature terminator analysis sheds light on a hidden world of bacterial transcriptional attenuation. Genome Biol. 2010, 11, R97. [Google Scholar] [CrossRef]
- Simons, R.W.; Houman, F.; Kleckner, N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 1987, 53, 85–96. [Google Scholar] [CrossRef]
- Powell, B.S.; Rivas, M.P.; Court, D.L.; Nakamura, Y.; Turnbough, C.L., Jr. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994, 22, 5765–5766. [Google Scholar] [CrossRef] [Green Version]
- Friedman, D.I.; Olson, E.R.; Johnson, L.L.; Alessi, D.; Craven, M.G. Transcription-dependent competition for a host factor: The function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990, 4, 2210–2222. [Google Scholar] [CrossRef]
- Amann, E.; Ochs, B.; Abel, K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 1988, 69, 301–315. [Google Scholar] [CrossRef]
- Saka, K.; Tadenuma, M.; Nakade, S.; Tanaka, N.; Sugawara, H.; Nishikawa, K.; Ichiyoshi, N.; Kitagawa, M.; Mori, H.; Osagawara, N.; et al. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 2005, 12, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.J.; Gross, C.A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 1988, 202, 45–58. [Google Scholar] [CrossRef]
- Poteete, A.R. Recombination phenotypes of Escherichia coli greA mutants. BMC Mol. Biol. 2011, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, F.; Rodriguez, R.L.; Greene, P.J.; Betlach, M.C.; Heyneker, H.L.; Boyer, H.W.; Crosa, J.H.; Falkow, S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 1977, 2, 95–113. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Moll, I.; Afonyushkin, T.; Vytvytska, O.; Kaberdin, V.R.; Bläsi, U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003, 9, 1308–1314. [Google Scholar] [CrossRef] [Green Version]
- Svetlov, V.; Artsimovitch, I. Purification of bacterial RNA polymerase: Tools and protocols. Methods Mol. Biol. 2015, 1276, 13–29. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dylewski, M.; Fernández-Coll, L.; Bruhn-Olszewska, B.; Balsalobre, C.; Potrykus, K. Autoregulation of greA Expression Relies on GraL Rather than on greA Promoter Region. Int. J. Mol. Sci. 2019, 20, 5224. https://doi.org/10.3390/ijms20205224
Dylewski M, Fernández-Coll L, Bruhn-Olszewska B, Balsalobre C, Potrykus K. Autoregulation of greA Expression Relies on GraL Rather than on greA Promoter Region. International Journal of Molecular Sciences. 2019; 20(20):5224. https://doi.org/10.3390/ijms20205224
Chicago/Turabian StyleDylewski, Maciej, Llorenç Fernández-Coll, Bożena Bruhn-Olszewska, Carlos Balsalobre, and Katarzyna Potrykus. 2019. "Autoregulation of greA Expression Relies on GraL Rather than on greA Promoter Region" International Journal of Molecular Sciences 20, no. 20: 5224. https://doi.org/10.3390/ijms20205224