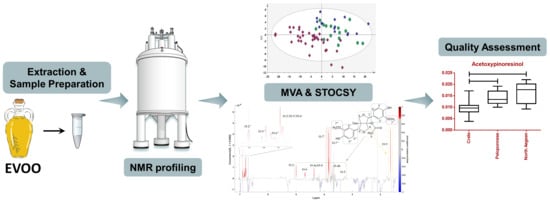
NMR-Based Metabolite Profiling and the Application of STOCSY toward the Quality and Authentication Assessment of European EVOOs
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sample Selection and Preparation
2.2. Acquisition, Processing, and Multivariate Analysis
2.2.1. European Samples
2.2.2. Greek Samples
2.3. Statistical Total Correlation Spectroscopy (STOCSY) and Biomarker Identification
2.4. Quality and Authentication Assessment
3. Materials and Methods
3.1. Chemical and Materials
3.1.1. Solvents and Reference Compounds
3.1.2. Sample Selection
3.1.3. Instrumentation
3.2. Sample Extraction
3.3. NMR Analysis
3.3.1. Sample Preparation
3.3.2. NMR Experimental Parameters
3.4. Computational Processing and Multivariate Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Angerosa, F. Sensory Quality of Olive Oils. In Handbook of Olive Oil: Analysis and Properties; Aparicio, R., Hardwood, J., Eds.; Springer: Gaithersburg, MD, USA, 2000; pp. 355–392. [Google Scholar]
- Frankel, E.N. Nutritional and Biological Properties of Extra Virgin Olive Oil. J. Agric. Food Chem. 2011, 59, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Olive Oil|European Commission. Available online: https://ec.europa.eu/info/food-farming-fisheries/plants-and-plant-products/plant-products/olive-oil_en (accessed on 8 September 2020).
- Vaclavik, L.; Cajka, T.; Hrbek, V.; Hajslova, J. Ambient Mass Spectrometry Employing Direct Analysis in Real Time (DART) Ion Source for Olive Oil Quality and Authenticity Assessment. Anal. Chim. Acta 2009, 645, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Codex Alimentarius Commission. Codex Alimentarius:Fats, Oils and Related Products, 2nd ed.; Food and Agriculture Orgnization of the United Nations (FAO): Rome, Italy, 2001. [Google Scholar]
- Commission Implementing Regulation (EU) No 1348/2013 of 16 December 2013 Amending Regulation (EEC) No 2568/91 on the Characteristics of Olive Oil and Olive-Residue Oil and on the Relevant Methods of Analysis; Publications Office of the European Union: Luxembourg. 2013. Available online: https://eur-lex.europa.eu/legal-content/GA/TXT/?uri=CELEX:32013R1348 (accessed on 9 February 2023).
- Commission Regulation (EU) No 61/2011 of 24 January 2011 Amending Regulation (EEC) No 2568/91 on the Characteristics of Olive Oil and Olive-Residue Oil and on the Relevant Methods of Analysis; Publications Office of the European Union: Luxembourg. 2011. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32011R0061 (accessed on 9 February 2023).
- Directive (EU) 2019/904 of the European Parliament and of the Council of 5 June 2019 on the Reduction of the Impact of Certain Plastic Products on the Environment; Publications Office of the European Union: Luxembourg. 2019. Available online: https://eur-lex.europa.eu/eli/dir/2019/904/oj (accessed on 9 February 2023).
- Commission Regulation (EC) No 1019/2002 of 13 June 2002 on Marketing Standards for Olive Oil; Publications Office of the European Union: Luxembourg. 2002. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32002R1019 (accessed on 9 February 2023).
- Quality Schemes Explained—European Commission. Available online: https://ec.europa.eu/info/food-farming-fisheries/food-safety-and-quality/certification/quality-labels/quality-schemes-explained_en (accessed on 5 September 2020).
- European Commission EAmbrosia—The EU Geographical Indications Register. Available online: https://ec.europa.eu/info/food-farming-fisheries/food-safety-and-quality/certification/quality-labels/geographical-indications-register/# (accessed on 10 September 2020).
- IOC STANDARDS, METHODS AND GUIDES—International Olive Council. Available online: https://www.internationaloliveoil.org/what-we-do/chemistry-standardisation-unit/standards-and-methods/ (accessed on 10 September 2020).
- D’Imperio, M.; Mannina, L.; Capitani, D.; Bidet, O.; Rossi, E.; Bucarelli, F.M.; Quaglia, G.B.; Segre, A. NMR and Statistical Study of Olive Oils from Lazio: A Geographical, Ecological and Agronomic Characterization. Food Chem. 2007, 105, 1256–1267. [Google Scholar] [CrossRef]
- Klikarová, J.; Rotondo, A.; Cacciola, F.; Česlová, L.; Dugo, P.; Mondello, L.; Rigano, F. The Phenolic Fraction of Italian Extra Virgin Olive Oils: Elucidation through Combined Liquid Chromatography and NMR Approaches. Food Anal. Methods 2019, 12, 1759–1770. [Google Scholar] [CrossRef]
- Ghisoni, S.; Lucini, L.; Angilletta, F.; Rocchetti, G.; Farinelli, D.; Tombesi, S.; Trevisan, M. Discrimination of Extra-Virgin-Olive Oils from Different Cultivars and Geographical Origins by Untargeted Metabolomics. Food Res. Int. 2019, 121, 746–753. [Google Scholar] [CrossRef]
- Petrakis, P.V.; Agiomyrgianaki, A.; Christophoridou, S.; Spyros, A.; Dais, P. Geographical Characterization of Greek Virgin Olive Oils (Cv. Koroneiki) Using 1H and 31P NMR Fingerprinting with Canonical Discriminant Analysis and Classification Binary Trees. J. Agric. Food Chem. 2008, 56, 3200–3207. [Google Scholar] [CrossRef]
- Mannina, L.; Fontanazza, G.; Patumi, M.; Ansanelli, G.; Segre, A. Italian and Argentine Olive Oils: A NMR and Gas Chromatographic Study. Grasas y Aceites 2001, 52, 380–388. [Google Scholar] [CrossRef]
- Olmo-García, L.; Wendt, K.; Kessler, N.; Bajoub, A.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Exploring the Capability of LC-MS and GC-MS Multi-Class Methods to Discriminate Virgin Olive Oils from Different Geographical Indications and to Identify Potential Origin Markers. Eur. J. Lipid Sci. Technol. 2019, 121, 1800336. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, S.; Liu, Y.; Song, H. Comparative Analysis of Volatiles of 15 Brands of Extra-Virgin Olive Oils Using Solid-Phase Micro-Extraction and Solvent-Assisted Flavor Evaporation. Molecules 2019, 24, 1512. [Google Scholar] [CrossRef] [PubMed]
- Da Ros, A.; Masuero, D.; Riccadonna, S.; Bubola, K.B.; Mulinacci, N.; Mattivi, F.; Lukić, I.; Vrhovsek, U. Complementary Untargeted and Targeted Metabolomics for Differentiation of Extra Virgin Olive Oils of Different Origin of Purchase Based on Volatile and Phenolic Composition and Sensory Quality. Molecules 2019, 24, 2896. [Google Scholar] [CrossRef] [Green Version]
- Girelli, C.R.; Del Coco, L.; Zelasco, S.; Salimonti, A.; Conforti, F.L.; Biagianti, A.; Barbini, D.; Fanizzi, F.P. Traceability of “Tuscan PGI” Extra Virgin Olive Oils by 1H NMR Metabolic Profiles Collection and Analysis. Metabolites 2018, 8, 60. [Google Scholar] [CrossRef]
- Vlahov, G.; Del Re, P.; Simone, N. Determination of Geographical Origin of Olive Oils Using 13C Nuclear Magnetic Resonance Spectroscopy. I—Classification of Olive Oils of the Puglia Region with Denomination of Protected Origin. J. Agric. Food Chem. 2003, 51, 5612–5615. [Google Scholar] [CrossRef]
- Mannina, L.; Dugo, G.; Salvo, F.; Cicero, L.; Ansanelli, G.; Calcagni, C.; Segre, A.L. Study of the Cultivar-Composition Relationship in Sicilian Olive Oils by GC, NMR, and Statistical Methods. J. Agric. Food Chem. 2003, 51, 120–127. [Google Scholar] [CrossRef]
- Culeddu, N.; Chessa, M.; Bandino, G.; Sedda, P.; Zurru, R.; Anedda, R.; Motroni, A.; Molinu, M.G.; Dettori, S.; Santona, M. Classification of Monovarietal Sardinian Extra Virgin Olive Oils by 1H NMR Metabolomic. Eur. J. Lipid Sci. Technol. 2017, 119, 1700035. [Google Scholar] [CrossRef]
- Dais, P.; Spyros, A. 31P NMR Spectroscopy in the Quality Control and Authentication of Extra-Virgin Olive Oil: A Review of Recent Progress. Magn. Reson. Chem. 2007, 45, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Kalogiouri, N.P.; Aalizadeh, R.; Thomaidis, N.S. Application of an Advanced and Wide Scope Non-Target Screening Workflow with LC-ESI-QTOF-MS and Chemometrics for the Classification of the Greek Olive Oil Varieties. Food Chem. 2018, 256, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Lioupi, A.; Nenadis, N.; Theodoridis, G. Virgin Olive Oil Metabolomics: A Review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1150, 122161. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, O.; Küchler, T. Reliable Classification of Olive Oil Origin Based on Minor Component Profile Using 1H-NMR and Multivariate Analysis. Eur. J. Lipid Sci. Technol. 2019, 121, 1900027. [Google Scholar] [CrossRef]
- Allard, P.M.; Genta-Jouve, G.; Wolfender, J.L. Deep Metabolome Annotation in Natural Products Research: Towards a Virtuous Cycle in Metabolite Identification. Curr. Opin. Chem. Biol. 2017, 36, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Computational Strategies for Metabolite Identification in Metabolomics. Bioanalysis 2009, 1, 1579–1596. [Google Scholar] [CrossRef]
- Sorokina, M.; Steinbeck, C. Review on Natural Products Databases: Where to Find Data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical Total Correlation Spectroscopy: An Exploratory Approach for Latent Biomarker Identification from Metabolic 1 H NMR Data Sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Ringele, G.B.L.; Beteinakis, S.; Papachristodoulou, A.; Axiotis, E.; Mikros, E.; Halabalaki, M. NMR Metabolite Profiling in the Quality and Authentication Assessment of Greek Honey—Exploitation of STOCSY for Markers Identification. Foods 2022, 11, 2853. [Google Scholar] [CrossRef] [PubMed]
- Beteinakis, S.; Papachristodoulou, A.; Gogou, G.; Katsikis, S.; Mikros, E.; Halabalaki, M. NMR-Based Metabolic Profiling of Edible Olives-Determination of Quality Parameters. Molecules 2020, 25, 3339. [Google Scholar] [CrossRef]
- Dais, P.; Hatzakis, E. Quality Assessment and Authentication of Virgin Olive Oil by NMR Spectroscopy: A Critical Review. Anal. Chim. Acta 2013, 765, 1–27. [Google Scholar] [CrossRef]
- Kosma, I.; Badeka, A.; Vatavali, K.; Kontakos, S.; Kontominas, M. Differentiation of Greek Extra Virgin Olive Oils According to Cultivar Based on Volatile Compound Analysis and Fatty Acid Composition. Eur. J. Lipid Sci. Technol. 2016, 118, 849–861. [Google Scholar] [CrossRef]
- Laincer, F.; Iaccarino, N.; Amato, J.; Pagano, B.; Pagano, A.; Tenore, G.; Tamendjari, A.; Rovellini, P.; Venturini, S.; Bellan, G.; et al. Characterization of Monovarietal Extra Virgin Olive Oils from the Province of Béjaïa (Algeria). Food Res. Int. 2016, 89, 1123–1133. [Google Scholar] [CrossRef]
- Olive Oil, Food Balance Sheets. Food and Agriculture Organization of the United Nations. FAOSTAT Statistical Database. 2014. Available online: https://www.fao.org/faostat/en/#search/olive%20oil (accessed on 10 July 2020).
- Anastasopoulos, E.; Kalogeropoulos, N.; Kaliora, A.C.; Kountouri, A.; Andrikopoulos, N.K. The Influence of Ripening and Crop Year on Quality Indices, Polyphenols, Terpenic Acids, Squalene, Fatty Acid Profile, and Sterols in Virgin Olive Oil (Koroneiki Cv.) Produced by Organic versus Non-Organic Cultivation Method. Int. J. Food Sci. Technol. 2011, 46, 170–178. [Google Scholar] [CrossRef]
- International Olive Council. Determination of Biophenols in Oive Oils by HPLC; International Olive Council: Madrid, Spain, 2017; pp. 1–8. [Google Scholar]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food Authentication: Techniques, Trends & Emerging Approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef]
- Nikou, T.; Witt, M.; Stathopoulos, P.; Barsch, A.; Halabalaki, M. Olive Oil Quality and Authenticity Assessment Aspects Employing FIA-MRMS and LC-Orbitrap MS Metabolomic Approaches. Front. Public Health 2020, 8, 558226. [Google Scholar] [CrossRef] [PubMed]
- Nikou, T.; Liaki, V.; Stathopoulos, P.; Sklirou, A.D.; Tsakiri, E.N.; Jakschitz, T.; Bonn, G.; Trougakos, I.P.; Halabalaki, M.; Skaltsounis, L.A. Comparison Survey of EVOO Polyphenols and Exploration of Healthy Aging-Promoting Properties of Oleocanthal and Oleacein. Food Chem. Toxicol. 2019, 125, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Boka, V.I.; Stathopoulou, K.; Benaki, D.; Gikas, E.; Aligiannis, N.; Mikros, E.; Skaltsounis, A.L. Could Multivariate Statistics Exploit HPTLC and NMR Data to Reveal Bioactive Compounds? The Case of Paeonia Mascula. Phytochem. Lett. 2017, 20, 379–385. [Google Scholar] [CrossRef]
- Yilmaz, A.; Nyberg, N.T.; Jaroszewski, J.W. Extraction of Alkaloids for NMR-Based Profiling: Exploratory Analysis of an Archaic Cinchona Bark Collection. Planta Med. 2012, 78, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Guldbrandsen, N.; Kostidis, S.; Schäfer, H.; De Mieri, M.; Spraul, M.; Skaltsounis, A.L.; Mikros, E.; Hamburger, M. NMR-Based Metabolomic Study on Isatis Tinctoria: Comparison of Different Accessions, Harvesting Dates, and the Effect of Repeated Harvesting. J. Nat. Prod. 2015, 78, 977–986. [Google Scholar] [CrossRef]
- Freire, R.T.; Bero, J.; Beaufay, C.; Selegato, D.M.; Coqueiro, A.; Choi, Y.H.; Quetin-Leclercq, J. Identification of Antiplasmodial Triterpenes from Keetia Species Using NMR-Based Metabolic Profiling. Metabolomics 2019, 15, 41. [Google Scholar] [CrossRef]
- Kew, W.; Goodall, I.; Uhrín, D. Analysis of Scotch Whisky by 1H NMR and Chemometrics Yields Insight into Its Complex Chemistry. Food Chem. 2019, 298, 125052. [Google Scholar] [CrossRef]
- Angelis, A.; Hamzaoui, M.; Aligiannis, N.; Nikou, T.; Michailidis, D.; Gerolimatos, P.; Termentzi, A.; Hubert, J.; Halabalaki, M.; Renault, J.H.; et al. An Integrated Process for the Recovery of High Added-Value Compounds from Olive Oil Using Solid Support Free Liquid-Liquid Extraction and Chromatography Techniques. J. Chromatogr. A 2017, 1491, 126–136. [Google Scholar] [CrossRef]
- Sarikaki, G.; Christoforidou, N.; Gaboriaud-Kolar, N.; Smith, A.B.; Kostakis, I.K.; Skaltsounis, A.L. Biomimetic Synthesis of Oleocanthal, Oleacein, and Their Analogues Starting from Oleuropein, a Major Compound of Olive Leaves. J. Nat. Prod. 2020, 83, 1735–1739. [Google Scholar] [CrossRef]
- Christophoridou, S.; Dais, P.; Tseng, L.I.H.; Spraul, M. Separation and Identification of Phenolic Compounds in Olive Oil by Coupling High-Performance Liquid Chromatography with Postcolumn Solid-Phase Extraction to Nuclear Magnetic Resonance Spectroscopy (LC-SPE-NMR). J. Agric. Food Chem. 2005, 53, 4667–4679. [Google Scholar] [CrossRef]
- Ruiz-Aracama, A.; Goicoechea, E.; Guillén, M.D. Direct Study of Minor Extra-Virgin Olive Oil Components without Any Sample Modification. 1H NMR Multisupression Experiment: A Powerful Tool. Food Chem. 2017, 228, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rodriguez, R.; Simonsen, U. Natural Triterpenoids from Olive Oil: Potential Activities against Cancer. In Natural Compounds as Inducers of Cell Death: Volume 1; Diederich, M., Noworyta, K., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 1, pp. 447–461. ISBN 9789400745759. [Google Scholar]
- Sokeng, A.J.T.; Sobolev, A.P.; Di Lorenzo, A.; Xiao, J.; Mannina, L.; Capitani, D.; Daglia, M. Metabolite Characterization of Powdered Fruits and Leaves from Adansonia Digitata L. (Baobab): A Multi-Methodological Approach. Food Chem. 2019, 272, 93–108. [Google Scholar] [CrossRef]
- Alonso-Salces, R.M.; Héberger, K.; Holland, M.V.; Moreno-Rojas, J.M.; Mariani, C.; Bellan, G.; Reniero, F.; Guillou, C. Multivariate Analysis of NMR Fingerprint of the Unsaponifiable Fraction of Virgin Olive Oils for Authentication Purposes. Food Chem. 2010, 118, 956–965. [Google Scholar] [CrossRef]
- Raffaele, S.; Francesco, A.; Livio, P. 1H and 13C NMR of Virgin Olive Oil. An Overview. Magn. Reson. Chem. 1997, 35, S133–S145. [Google Scholar] [CrossRef]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High Resolution NMR Spectroscopy as a Structural and Analytical Tool for Unsaturated Lipids in Solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef]
- International Olive Council COI/T.20/DOC.15/2015. Available online: http://www.internationaloliveoil.org/estaticos/view/224-testing-methods (accessed on 15 October 2020).
- López-Biedma, A.; Sánchez-Quesada, C.; Delgado-Rodríguez, M.; Gaforio, J.J. The Biological Activities of Natural Lignans from Olives and Virgin Olive Oils: A Review. J. Funct. Foods 2016, 26, 36–47. [Google Scholar] [CrossRef]
- Kanakis, P.; Termentzi, A.; Michel, T.; Gikas, E.; Halabalaki, M.; Skaltsounis, A.L. From Olive Drupes to Olive Oil. An HPLC-Orbitrap-Based Qualitative and Quantitative Exploration of Olive Key Metabolites. Planta Med. 2013, 79, 1576–1587. [Google Scholar] [CrossRef]
- Ayeleso, T.B. Oleanolic Acid and Its Derivatives: Biological Chronic Diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef]
- Lozano-Mena, G.; Sánchez-González, M.; Juan, M.E.; Planas, J.M. Maslinic Acid, a Natural Phytoalexin-Type Triterpene from Olives—A Promising Nutraceutical? Molecules 2014, 19, 11538–11559. [Google Scholar] [CrossRef]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.P.; Capitani, D. NMR and Chemometrics in Tracing European Olive Oils: The Case Study of Ligurian Samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef]
- Mohamed, M.B.; Rocchetti, G.; Montesano, D.; Ali, S.B.; Guasmi, F.; Grati-Kamoun, N.; Lucini, L. Discrimination of Tunisian and Italian Extra-Virgin Olive Oils According to Their Phenolic and Sterolic Fingerprints. Food Res. Int. 2018, 106, 920–927. [Google Scholar] [CrossRef]
- Ün, İ.; Ok, S. Analysis of Olive Oil for Authentication and Shelf Life Determination. J. Food Sci. Technol. 2018, 55, 2476–2487. [Google Scholar] [CrossRef] [PubMed]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. Icoshift: A Versatile Tool for the Rapid Alignment of 1D NMR Spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beteinakis, S.; Papachristodoulou, A.; Kolb, P.; Rösch, P.; Schwarzinger, S.; Mikros, E.; Halabalaki, M. NMR-Based Metabolite Profiling and the Application of STOCSY toward the Quality and Authentication Assessment of European EVOOs. Molecules 2023, 28, 1738. https://doi.org/10.3390/molecules28041738
Beteinakis S, Papachristodoulou A, Kolb P, Rösch P, Schwarzinger S, Mikros E, Halabalaki M. NMR-Based Metabolite Profiling and the Application of STOCSY toward the Quality and Authentication Assessment of European EVOOs. Molecules. 2023; 28(4):1738. https://doi.org/10.3390/molecules28041738
Chicago/Turabian StyleBeteinakis, Stavros, Anastasia Papachristodoulou, Peter Kolb, Paul Rösch, Stephan Schwarzinger, Emmanuel Mikros, and Maria Halabalaki. 2023. "NMR-Based Metabolite Profiling and the Application of STOCSY toward the Quality and Authentication Assessment of European EVOOs" Molecules 28, no. 4: 1738. https://doi.org/10.3390/molecules28041738