A Novel Kinetic Modeling of Enzymatic Hydrolysis of Sugarcane Bagasse Pretreated by Hydrothermal and Organosolv Processes
Abstract
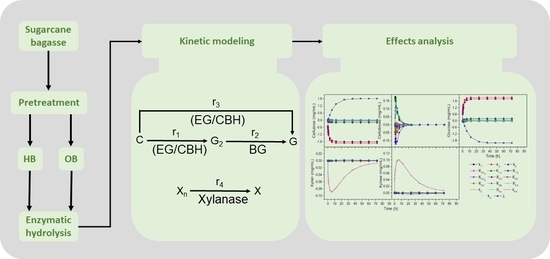
:1. Introduction
2. Results and Discussion
2.1. Estimation of Kinetic Parameters for Sugarcane Bagasse under Hydrothermal and Organosolv Pretreatment
2.2. Plackett–Burman Design for the Selection of Significant Parameters
3. Materials and Methods
3.1. Biomass Preparation and Pretreatment
3.2. Enzymatic Activity
3.3. Enzymatic Hydrolysis
3.4. Quantification of Sugars
3.5. Kinetic Model
- (1)
- The enzymatic adsorption follows the Langmuir adsorption isotherm, where the r1 and r3 reactions occur on the cellulose surface;
- (2)
- Enzymatic deactivation by thermal and mechanical effects was negligible;
- (3)
- The cellulosic matrix was uniform in terms of enzyme accessibility in the substrate, without distinction between the amorphous and crystalline fractions of cellulose;
- (4)
- The cellulose consisted of EG, CBH, and low β-glucosidase activity. The model did not distinguish EG from CBH. Due to the low amount of β-glucosidase, the model considered the enzyme only from Aspergillus niger;
- (5)
- The xylanase from the reaction medium was present in the cellulase used in the experimental assays;
- (6)
- The hemicelluloses of HB and OB were composed solely of xylan;
- (7)
- The conversion of cellobiose into glucose represented by r2 occurred in solution and followed the Michaelis–Menten kinetics;
- (8)
- The conversion of xylan into xylose represented by r4 occurred in a single reaction, absent intermediate compounds such as xylobiose;
- (9)
- The proportion of lignin exposed to the enzyme of the total lignin present in the pretreated bagasse was equal to 1, demonstrating that cellulose did not block the adsorption of enzymes on lignin [24];
- (10)
- β-glucosidase did not adsorb to cellulose and lignin;
- (11)
- The lignin of the pretreated biomass was not degraded during enzymatic hydrolysis.
3.6. Parameter Estimation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Qi, F.; Wright, M. A novel optimization approach to estimating kinetic parameters of the enzymatic hydrolysis of corn stover. AIMS Energy 2016, 4, 52–67. [Google Scholar] [CrossRef]
- Preethi, M.G.; Kumar, G.; Karthikeyan, O.P.; Varjani, S.; Banu, J.R. Lignocellulosic biomass as an optimistic feedstock for the production of biofuels as valuable energy source: Techno-economic analysis, Environmental Impact Analysis, Breakthrough and Perspectives. Environ. Technol. Innov. 2021, 24, 102080. [Google Scholar] [CrossRef]
- Solarte-Toro, J.C.; Chacón-Pérez, Y.; Cardona-Alzate, C.A. Evaluation of biogas and syngas as energy vectors for heat and power generation using lignocellulosic biomass as raw material. Electron. J. Biotechnol. 2018, 33, 52–62. [Google Scholar] [CrossRef]
- Gao, W.; Li, Z.; Liu, T.; Wang, Y. Production of high-concentration fermentable sugars from lignocellulosic biomass by using high solids fed-batch enzymatic hydrolysis. Biochem. Eng. J. 2021, 176, 108186. [Google Scholar] [CrossRef]
- Suarez, C.A.G.; Cavalcanti-Montaño, I.D.; da Costa Marques, R.G.; Furlan, F.F.; da Mota e Aquino, P.L.; de Campos Giordano, R.; de Sousa, R. Modeling the Kinetics of Complex Systems: Enzymatic Hydrolysis of Lignocellulosic Substrates. Appl. Biochem. Biotechnol. 2014, 173, 1083–1096. [Google Scholar] [CrossRef]
- Ashraf, M.T.; Thomsen, M.; Schmidt, J. Hydrothermal Pretreatment and Enzymatic Hydrolysis of Mixed Green and Woody Lignocellulosics from Arid Regions. Bioresour. Technol. 2017, 238, 369–378. [Google Scholar] [CrossRef]
- Lee, H.V.; Hamid, S.B.A.; Zain, S.K. Conversion of Lignocellulosic Biomass to Nanocellulose: Structure and Chemical Process. Sci. World J. 2014, 2014, 631013. [Google Scholar] [CrossRef] [Green Version]
- Keller, R.G.; Weyand, J.; Vennekoetter, J.B.; Kamp, J.; Wessling, M. An electro-Fenton process coupled with nanofiltration for enhanced conversion of cellobiose to glucose. Catal. Today 2021, 364, 230–241. [Google Scholar] [CrossRef]
- Nadar, D.; Naicker, K.; Lokhat, D. Ultrasonically-Assisted Dissolution of Sugarcane Bagasse during Dilute Acid Pretreatment: Experiments and Kinetic Modeling. Energies 2020, 13, 5627. [Google Scholar] [CrossRef]
- Mpatani, F.M.; Aryee, A.A.; Kani, A.N.; Wen, K.; Dovi, E.; Qu, L.; Li, Z.; Han, R. Removal of methylene blue from aqueous medium by citrate modified bagasse: Kinetic, Equilibrium and Thermodynamic study. Bioresour. Technol. Rep. 2020, 11, 100463. [Google Scholar] [CrossRef]
- Silveira, M.H.L.; Chandel, A.K.; Vanelli, B.A.; Sacilotto, K.S.; Cardoso, E.B. Production of hemicellulosic sugars from sugarcane bagasse via steam explosion employing industrially feasible conditions: Pilot scale study. Bioresour. Technol. Rep. 2018, 3, 138–146. [Google Scholar] [CrossRef]
- Nikodinovic-Runic, J.; Guzik, M.; Kenny, S.T.; Babu, R.; Werker, A.; Connor, K.E.O. Chapter Four-Carbon-Rich Wastes as Feedstocks for Biodegradable Polymer (Polyhydroxyalkanoate) Production Using Bacteria. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: New York, NY, USA, 2013; Volume 84, pp. 139–200. [Google Scholar]
- da Siva Martins, L.H.; Komesu, A.; Moreira Neto, J.; de Oliveira, J.A.R.; Rabelo, S.C.; da Costa, A.C. Pretreatment of sugarcane bagasse by OX-B to enhancing the enzymatic hydrolysis for ethanol fermentation. J. Food Process Eng. 2021, 44, e13579. [Google Scholar] [CrossRef]
- Sun, S.; Sun, S.; Cao, X.; Sun, R. The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour. Technol. 2016, 199, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Peng, X.; Sun, S.; Zhong, L.; Sun, R. Hydrothermal Conversion of Bamboo: Identification and Distribution of the Components in Solid Residue, Water-Soluble and Acetone-Soluble Fractions. J. Agric. Food Chem. 2014, 62, 12360–12365. [Google Scholar] [CrossRef]
- Sun, S.; Cao, X.; Sun, S.; Xu, F.; Song, X.; Sun, R.-C.; Jones, G.L. Improving the enzymatic hydrolysis of thermo-mechanical fiber from Eucalyptus urophylla by a combination of hydrothermal pretreatment and alkali fractionation. Biotechnol. Biofuels 2014, 7, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Villota, D.S.; Acosta-Pavas, J.C.; Betancur-Ramírez, K.J.; Ruiz-Colorado, A.A. Modeling and Kinetic Parameter Estimation of the Enzymatic Hydrolysis Process of Lignocellulosic Materials for Glucose Production. Ind. Eng. Chem. Res. 2020, 59, 16851–16867. [Google Scholar] [CrossRef]
- Huron, M.; Hudebine, D.; Lopes Ferreira, N.; Lachenal, D. Mechanistic modeling of enzymatic hydrolysis of cellulose integrating substrate morphology and cocktail composition. Biotechnol. Bioeng. 2016, 113, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.P.; Lynd, L.R. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotechnol. Bioeng. 2004, 88, 797–824. [Google Scholar] [CrossRef]
- Godoy, C.M.d.; Machado, D.L.; Costa, A.C.d. Batch and fed-batch enzymatic hydrolysis of pretreated sugarcane bagasse–Assays and modeling. Fuel 2019, 253, 392–399. [Google Scholar] [CrossRef]
- Ponce, G.H.S.F.; Moreira Neto, J.; De Jesus, S.S.; Miranda, J.C.d.C.; Maciel Filho, R.; Andrade, R.R.d.; Wolf Maciel, M.R. Sugarcane molasses fermentation with in situ gas stripping using low and moderate sugar concentrations for ethanol production: Experimental data and modeling. Biochem. Eng. J. 2016, 110, 152–161. [Google Scholar] [CrossRef]
- Neto, J.M.; dos Reis Garcia, D.; Rueda, S.M.G.; da Costa, A.C. Study of kinetic parameters in a mechanistic model for enzymatic hydrolysis of sugarcane bagasse subjected to different pretreatments. Bioprocess Biosyst. Eng. 2013, 36, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, P.; Knapp, B. User’s Guide to Pikaia 1.0-Ncar Technical Note 418+IA; National Center for Atmospheric Research: Boulder, CO, USA, 1995. [Google Scholar]
- Zheng, Y.; Pan, Z.; Zhang, R.; Jenkins, B. Kinetic Modeling for Enzymatic Hydrolysis of Pretreated Creeping Wild Ryegrass. Biotechnol. Bioeng. 2009, 102, 1558–1569. [Google Scholar] [CrossRef]
- Khodaverdi, M.; Jeihanipour, A.; Karimi, K.; Taherzadeh, M.J. Kinetic modeling of rapid enzymatic hydrolysis of crystalline cellulose after pretreatment by NMMO. J. Ind. Microbiol. Biotechnol. 2012, 39, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Angarita, J.D.; Souza, R.B.A.; Cruz, A.J.G.; Biscaia, E.C.; Secchi, A.R. Kinetic modeling for enzymatic hydrolysis of pretreated sugarcane straw. Biochem. Eng. J. 2015, 104, 10–19. [Google Scholar] [CrossRef]
- Flores-Sánchez, A.; Flores-Tlacuahuac, A.; Pedraza-Segura, L.L. Model-Based Experimental Design to Estimate Kinetic Parameters of the Enzymatic Hydrolysis of Lignocellulose. Ind. Eng. Chem. Res. 2013, 52, 4834–4850. [Google Scholar] [CrossRef]
- Prunescu, R.M.; Sin, G. Dynamic modeling and validation of a lignocellulosic enzymatic hydrolysis process–A demonstration scale study. Bioresour. Technol. 2013, 150, 393–403. [Google Scholar] [CrossRef]
- Kadam, K.L.; Rydholm, E.C.; McMillan, J.D. Development and Validation of a Kinetic Model for Enzymatic Saccharification of Lignocellulosic Biomass. Biotechnol. Prog. 2004, 20, 698–705. [Google Scholar] [CrossRef]
- Scott, F.; Li, M.; Williams, D.L.; Conejeros, R.; Hodge, D.B.; Aroca, G. Corn stover semi-mechanistic enzymatic hydrolysis model with tight parameter confidence intervals for model-based process design and optimization. Bioresour. Technol. 2015, 177, 255–265. [Google Scholar] [CrossRef]
- Morales-Rodriguez, R.; Meyer, A.S.; Gernaey, K.V.; Sin, G. Dynamic model-based evaluation of process configurations for integrated operation of hydrolysis and co-fermentation for bioethanol production from lignocellulose. Bioresour. Technol. 2011, 102, 1174–1184. [Google Scholar] [CrossRef]
- Morales-Rodriguez, R.; Meyer, A.S.; Gernaey, K.V.; Sin, G. A framework for model-based optimization of bioprocesses under uncertainty: Lignocellulosic ethanol production case. Comput. Chem. Eng. 2012, 42, 115–129. [Google Scholar] [CrossRef] [Green Version]
- Machado, D.L.; Moreira Neto, J.; da Cruz Pradella, J.G.; Bonomi, A.; Rabelo, S.C.; da Costa, A.C. Adsorption characteristics of cellulase and β-glucosidase on Avicel, pretreated sugarcane bagasse, and lignin. Biotechnol. Appl. Biochem. 2015, 62, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Qing, Q.; Yang, B.; Wyman, C.E. Xylooligomers are strong inhibitors of cellulose hydrolysis by enzymes. Bioresour. Technol. 2010, 101, 9624–9630. [Google Scholar] [CrossRef] [PubMed]
- Moreira Neto, J.; Machado, D.L.; Bonomi, A.; Gonçalves, V.O.O.; Martins, L.H.S.; Costa, J.M.; Costa, A.C. Cellulase adsorption on pretreated sugarcane bagasse during enzymatic hydrolysis. Sugar Tech. 2023. [Google Scholar]
- Drissen, R.E.T.; Maas, R.H.W.; Van Der Maarel, M.J.E.C.; Kabel, M.A.; Schols, H.A.; Tramper, J.; Beeftink, H.H. A generic model for glucose production from various cellulose sources by a commercial cellulase complex. Biocatal. Biotransform. 2007, 25, 419–429. [Google Scholar] [CrossRef]





| Pretreatment | Bagasse Concentration % (m/v) | Cellulose (mg/mL) | Lignin (mg/mL) | Xylan (mg/mL) |
|---|---|---|---|---|
| HB | 4 | 24.4 ± 3.9 | 12.79 ± 0.02 | 0.84 ± 0.02 |
| HB | 6 | 36.6 ± 5.8 | 19.18 ± 0.03 | 1.26 ± 0.04 |
| HB | 8 | 48.9 ± 7.8 | 25.58 ± 0.04 | 1.68 ± 0.05 |
| HB | 10 | 61.1 ± 9.7 | 31.97 ± 0.05 | 2.10 ± 0.06 |
| HB | 12 | 73.3 ± 11.6 | 38.36 ± 0.06 | 2.52 ± 0.07 |
| OB | 4 | 34.8 ± 1.6 | 2.7 ± 1.1 | 1.8 ± 0.1 |
| OB | 6 | 52.2 ± 2.4 | 4 ± 2 | 2.65 ± 0.15 |
| OB | 8 | 69.5 ± 3.2 | 5.3 ± 2.2 | 3.5 ± 0.2 |
| OB | 10 | 87 ± 4 | 6.6 ± 2.7 | 4.42 ± 0.25 |
| OB | 12 | 104.3 ± 4.8 | 8 ± 3 | 5.3 ± 0.3 |
| Bagasse Concentration % (m/v) | EG/CBH (mg/mL) | BG (mg/mL) | Xylanase (U/mL) |
|---|---|---|---|
| 4 | 0.420 | 0.1017 | 5.853 |
| 6 | 0.629 | 0.1526 | 8.779 |
| 8 | 0.839 | 0.2034 | 11.706 |
| 10 | 1.049 | 0.2543 | 14.632 |
| 12 | 1.259 | 0.3052 | 17.559 |
| Current Study | [24] | [25] | [26] | [27] | [28] | ||
|---|---|---|---|---|---|---|---|
| Parameter | HB | OB | Acid Treatment—Wild Ryegrass | Cotton Pretreated with N-Oxide-N-Methylmorpholine | Sugarcane Straw | Corncob Stock | Wheat Straw |
| k1r (mL/mg h) | 19.178 | 20.289 | 16.5 | 32.10 | 0.509 | 94.72 | 1.224 |
| k2r (h−1) | 196.56 | 230.82 | 267.6 | 263.89 | 165.7 | 432.16 | 252 |
| k3r (mL/mg h) | 8.576 | 7.236 | 7.1 | 13.56 | 12.75 | 958.3 | 19.08 |
| K1iG2 (mg/mL) | 0.769 | 0.11 | 0.02 | 7.52 | 0.016 | 1.00 × 10−5 | 0.0014 |
| K1iG (mg/mL) | 0.03 | 4.875 | 0.1 | 0.34 | 0.710 | 7.33 | 0.073 |
| K1iX (mg/mL) | 31.92 | 285.15 | - | - | 0.559 | 8.92 | 0.1007 |
| K2iG (mg/mL) | 14.853 | 10.02 | 2.1 | 3.19 | 0.011 | 1.45 × 10−5 | 3.9 |
| K2m (mg/mL) | 22.48 | 11.295 | 25.5 | 11.63 | 47.20 | 0.022 | 24.3 |
| K2iX (mg/mL) | 278.2 | 51.48 | - | - | 110.0 | 39.19 | 201 |
| K3iG2 (mg/mL) | 0.913 | 2.574 | 132.5 | 38.41 | 89.18 | 7.33 | 132 |
| K3iG (mg/mL) | 0.853 | 0.167 | 0.01 | 1.58 | 0.551 | 1.15 × 10−3 | 0.34 |
| K3iX (mg/mL) | 86.38 | 180.22 | - | - | 0.581 | 6.13 | 0.029 |
| k4 (h−1) | 18.066 | 37.56 | - | - | 13.46 b | 167.27 b | 9.72 b |
| Keq (U/mL) | 0.0786 | 0.0066 | - | - | - | - | - |
| K4iX (mg/mL) | 0.0111 | 0.0262 | - | - | 134.1 | 23.12 | 201 |
| ks (mg/mL) | 0.0354 | 45.8 | - | - | - | - | - |
| a λ (h−1) | 0.2004 | - | - | - | - | - | |
| Parameter | Cellulose | Cellobiose | Glucose | Xylan | Xylose |
|---|---|---|---|---|---|
| k1r | |||||
| k2r | |||||
| k3r | |||||
| K1iG2 | |||||
| K1iG | |||||
| K1iX | |||||
| K2iG | |||||
| K2m | |||||
| K2iX | |||||
| K3iG2 | |||||
| K3iG | |||||
| K3iX | |||||
| k4 | |||||
| Keq | |||||
| K4iX | |||||
| k4s | |||||
| λ |
| Parameters | a | b | C | R2 |
|---|---|---|---|---|
| Emax (HB) | 3.607 | −0.00719 | −0.0000772 | 0.980 |
| Kp (HB) | 0.2501 | 0.00134 | −0.000146 | 0.979 |
| Emax (OB) | 3.383 | −0.0027 | 0.000003 | 0.996 |
| Kp (OB) | 1.008 | −0.014 | 0.00008 | 0.889 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira Neto, J.; Costa, J.M.; Bonomi, A.; Costa, A.C. A Novel Kinetic Modeling of Enzymatic Hydrolysis of Sugarcane Bagasse Pretreated by Hydrothermal and Organosolv Processes. Molecules 2023, 28, 5617. https://doi.org/10.3390/molecules28145617
Moreira Neto J, Costa JM, Bonomi A, Costa AC. A Novel Kinetic Modeling of Enzymatic Hydrolysis of Sugarcane Bagasse Pretreated by Hydrothermal and Organosolv Processes. Molecules. 2023; 28(14):5617. https://doi.org/10.3390/molecules28145617
Chicago/Turabian StyleMoreira Neto, João, Josiel Martins Costa, Antonio Bonomi, and Aline Carvalho Costa. 2023. "A Novel Kinetic Modeling of Enzymatic Hydrolysis of Sugarcane Bagasse Pretreated by Hydrothermal and Organosolv Processes" Molecules 28, no. 14: 5617. https://doi.org/10.3390/molecules28145617