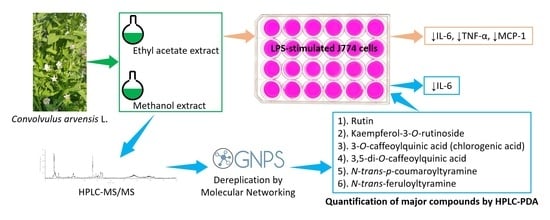
Dereplication and Quantification of Major Compounds of Convolvulus arvensis L. Extracts and Assessment of Their Effect on LPS-Activated J774 Macrophages
Abstract
:1. Introduction
2. Results and Discussion
2.1. Extraction Yield and MTT Assay of Crude Extracts
2.2. Effect of ARE, ARM, and ARW on the Expression of Pro-Inflammatory Mediators in LPS-Stimulated J744 Cells
2.3. Dereplication of ARE and ARM
2.4. Quantification and Biological Screening of Major Compounds of ARE and ARM
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Collection of Plant
3.3. Preparation of Crude Extracts
3.4. Cell Cultures
3.5. MTT Assay
3.6. Effect of Crude Extracts and Identified Major Compounds on the Expression of Pro-Inflammatory Mediators in LPS-Stimulated J774 Cells
3.7. Real-Time Quantitative PCR (qPCR)
3.8. Cytokines Quantification by ELISA
3.9. HPLC-PDA Analysis
3.10. HPLC-DAD-HRMS/MS Analysis
3.11. MS Data Treatment
3.12. Mass Spectral Organization and Dereplication
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Uttra, A.M.; Ahsan, H.; Hasan, U.H.; Chaudhary, M.A. Traditional Medicines of Plant Origin Used for the Treatment of Inflammatory Disorders in Pakistan: A Review. J. Tradit. Chin. Med. 2018, 38, 636–656. [Google Scholar] [CrossRef]
- Kaur, M.; Kalia, A.N. Convolvulus Arvensis—A Useful Weed. Int. J. Pharm. Pharm. Sci. 2012, 4, 38–40. [Google Scholar]
- Iqbal, H.; Sher, Z.; Khan, Z.U. Medicinal Plants from Salt Range Pind Dadan Khan, District Jhelum, Punjab, Pakistan. J. Med. Plant. Res. 2011, 5, 2157–2168. [Google Scholar]
- Ijaz, F.; Iqbal, Z.; Alam, J.; Khan, S.M.; Afzal, A.; Rahman, I.U.; Afzal, M.; Islam, M. Ethno Medicinal Study upon Folk Recipes against Various Human Diseases in Sarban Hills, Abbottabad, Pakistan. WJZ 2015, 10, 41–46. [Google Scholar] [CrossRef]
- Ahmed, N.; Mahmood, A.; Mahmood, A.; Tahir, S.S.; Bano, A.; Malik, R.N.; Hassan, S.; Ishtiaq, M. Relative Importance of Indigenous Medicinal Plants from Layyah District, Punjab Province, Pakistan. J. Ethnopharmacol. 2014, 155, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.S.; Roy, S.D.; Ghaffari, M.A.; Uzair, M.; Ijaz, A.S.; Khan, T.R. Survey of Ethno-Medicinal Weeds of District Rajhan Pur, Punjab, Pakistan. Ind. Res. J. Pharm. Sci. 2014, 1, 38–45. [Google Scholar]
- Abbas, Z.; Khan, S.M.; Abbasi, A.M.; Pieroni, A.; Ullah, Z.; Iqbal, M.; Ahmad, Z. Ethnobotany of the Balti Community, Tormik Valley, Karakorum Range, Baltistan, Pakistan. J. Ethnobiol. Ethnomed. 2016, 12, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Elzaawely, A.A.; Tawata, S. Antioxidant Activity of Phenolic Rich Fraction Obtained from Convolvulus arvensis L. Leaves Grown in Egypt. Asian J. Crop. Sci. 2012, 4, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Khakimov, Z.; Rakhmanov, A.K.; Yakubova, U.B.; Shukurlaev, K.S. Experimental Substantiation of Anti-Inflammatory Activity of a Gel Containing Convolvulus arvensis Extract in Carrageenan-Induced Aseptic Arthritis. Natl. J. Physiol. Pharm. Pharmacol. 2021, 11, 645–648. [Google Scholar] [CrossRef]
- Saleem, U.; Zaib, S.; Khalid, S.; Anwar, F.; Akhtar, M.F.; Ahmad, B. Chemical Characterization, Docking Studies, Anti-Arthritic Activity and Acute Oral Toxicity of Convolvulus arvensis L. Leaves. Asian Pac. J. Trop. Biomed. 2020, 10, 442–451. [Google Scholar] [CrossRef]
- Ali, M.; Qadir, M.I.; Saleem, M.; Janbaz, K.H.; Gul, H.; Hussain, L.; Ahmad, B. Hepatoprotective Potential of Convolvulus arvensis against Paracetamol-Induced Hepatotoxicity. Bangladesh J. Pharmacol. 2013, 8, 300–304. [Google Scholar] [CrossRef] [Green Version]
- Awaad, A.S.; Mohamed, N.H.; El-Sayed, N.H. Phenolics of Convolvulus arvensis L. and Their Related Pharmacological Activity. Asian J. Chem. 2006, 18, 2818–2826. [Google Scholar]
- Todd, F. Tropane Alkaloids and Toxicity of Convolvulus arvensis. Phytochemistry 1995, 39, 301–303. [Google Scholar] [CrossRef]
- Fan, B.-Y.; He, Y.; Lu, Y.; Yang, M.; Zhu, Q.; Chen, G.-T.; Li, J.-L. Glycosidic Acids with Unusual Aglycone Units from Convolvulus arvensis. J. Nat. Prod. 2019, 82, 1593–1598. [Google Scholar] [CrossRef]
- Muhsinah, A.B.; Alsayari, A.; Thajudeen, K.Y.; Asiri, Y.I.; Nanjaian, M. Simultaneous Estimation of Rutin and Quercetin in Bidens pilosa, Convolvulus arvensis and Neurada procumbens by RP-HPLC. Res. J. Pharm. Technol. 2020, 13, 3305–3310. [Google Scholar] [CrossRef]
- Sowemimo, B.O.; Farnsworth, N.R. Phytochemical Investigation of Convolvulus arvensis (Convolvulaceae). J. Pharm. Sc. 1973, 62, 678–679. [Google Scholar] [CrossRef]
- Fujiwara, N.; Kobayashi, K. Macrophages in Inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 281–286. [Google Scholar] [CrossRef]
- DuBois, R.N.; Abramson, S.B.; Crofford, L.; Gupta, R.A.; Simon, L.S.; van de Putte, L.B.A.; Lipsky, P.E. Cyclooxygenase in Biology and Disease. FASEB J. 1998, 12, 1063–1073. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, R.; Lahti, A.; Kankaanranta, H.; Moilanen, E. Nitric Oxide Production and Signaling in Inflammation. CDTIA 2005, 4, 471–479. [Google Scholar] [CrossRef]
- Netea, M.G.; Balkwill, F.; Chonchol, M.; Cominelli, F.; Donath, M.Y.; Giamarellos-Bourboulis, E.J.; Golenbock, D.; Gresnigt, M.S.; Heneka, M.T.; Hoffman, H.M.; et al. A Guiding Map for Inflammation. Nat. Immunol. 2017, 18, 826–831. [Google Scholar] [CrossRef] [Green Version]
- Tasneem, S.; Liu, B.; Li, B.; Choudhary, M.I.; Wang, W. Molecular Pharmacology of Inflammation: Medicinal Plants as Anti-Inflammatory Agents. Pharmacol. Res. 2019, 139, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, A.; Kishimoto, T. IL-6: Regulator of Treg/Th17 Balance. Eur. J. Immunol. 2010, 40, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Cytokines in Inflammatory Bowel Disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef]
- Yoshimura, T. The Production of Monocyte Chemoattractant Protein-1 (MCP-1)/CCL2 in Tumor Microenvironments. Cytokine 2017, 98, 71–78. [Google Scholar] [CrossRef]
- Umamaheswaran, S.; Dasari, S.K.; Yang, P.; Lutgendorf, S.K.; Sood, A.K. Stress, Inflammation, and Eicosanoids: An Emerging Perspective. Cancer Metastasis Rev. 2018, 37, 203–211. [Google Scholar] [CrossRef]
- Salehi, B.; Krochmal-Marczak, B.; Skiba, D.; Patra, J.K.; Das, S.K.; Das, G.; Popović-Djordjević, J.B.; Kostić, A.Ž.; Anil Kumar, N.V.; Tripathi, A.; et al. Convolvulus Plant—A Comprehensive Review from Phytochemical Composition to Pharmacy. Phytother. Res. 2020, 34, 315–328. [Google Scholar] [CrossRef]
- Ambriz-Pérez, D.L.; Leyva-López, N.; Gutierrez-Grijalva, E.P.; Heredia, J.B. Phenolic Compounds: Natural Alternative in Inflammation Treatment. A Review. Cogent Food Agric. 2016, 2, 1131412. [Google Scholar] [CrossRef]
- Clifford, M.N.; Knight, S.; Kuhnert, N. Discriminating between the Six Isomers of Dicaffeoylquinic Acid by LC-MSn. J. Agric. Food Chem. 2005, 53, 3821–3832. [Google Scholar] [CrossRef]
- Ablajan, K.; Abliz, Z.; Shang, X.-Y.; He, J.-M.; Zhang, R.-P.; Shi, J.-G. Structural Characterization of Flavonol 3,7-Di-O-Glycosides and Determination of the Glycosylation Position by Using Negative Ion Electrospray Ionization Tandem Mass Spectrometry. J. Mass Spectrom. 2006, 41, 352–360. [Google Scholar] [CrossRef]
- Hrichi, S.; Chaabane-Banaoues, R.; Giuffrida, D.; Mangraviti, D.; Oulad el Majdoub, Y.; Rigano, F.; Mondello, L.; Babba, H.; Mighri, Z.; Cacciola, F. Effect of Seasonal Variation on the Chemical Composition and Antioxidant and Antifungal Activities of Convolvulus althaeoides L. Leaf Extracts. Arab. J. Chem. 2020, 13, 5651–5668. [Google Scholar] [CrossRef]
- Hassine, M.; Zardi-Berguaoui, A.; Harzallah-Skhiri, F.; Abreu, P.; Jannet, H.B.; Hamza, M.A. Isolation and Structure Elucidation of Secondary Metabolites from the Roots of the Tunisian Convolvulus dorycnium. Chem. Nat. Compd. 2016, 52, 830–833. [Google Scholar] [CrossRef]
- Krzaczek, T.; Gogucka-Kocka, A.; Ryn, D. Chromatographical Analysis of Phenolic Compounds in Herb Convolvulus arvensis L. Herba Pol. 2004, 50, 17–22. [Google Scholar]
- Yaoya, S.; Kanho, H.; Mikami, Y.; Itani, T.; Umehara, K.; Kuroyanagi, M. Umbelliferone Released from Hairy Root Cultures of Pharbitis nil Treated with Copper Sulfate and Its Subsequent Glucosylation. Biosci. Biotechnol. Biochem. 2004, 68, 1837–1841. [Google Scholar] [CrossRef]
- Takenaka, M.; Nanayama, K.; Isobe, S.; Murata, M. Changes in Caffeic Acid Derivatives in Sweet Potato (Ipomoea batatas L.) during Cooking and Processing. Biosci. Biotechnol. Biochem. 2006, 70, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-L.; Hua, Z.; Zhao, B.-Y.; Tang, W.-X.; Zhang, S.-J. Studies on the chemical constituents of Pharbitis purpurea. Zhong Yao Cai 2010, 33, 1571–1574. [Google Scholar]
- Belaouira, R.; Marchioni, E.; Benayache, F.; Benayache, S. On-Line Screening and Identification of Polyphenolic Antioxidant Compounds of Convolvulus trabutianus. Nat. Prod. Res. 2020, 34, 1490–1493. [Google Scholar] [CrossRef]
- Wang, W.; Xuan, L. New Acyclic Sesquiterpenoid Derivatives and a Monoterpene Disaccharide from Dichondra repens Forst. Phytochem. Lett. 2015, 14, 23–26. [Google Scholar] [CrossRef]
- Shahat, A.A.; Abdel-Azim, N.S.; Pieters, L.; Vlietinck, A.J. Flavonoids from Cressa cretica. Pharm. Biol. 2004, 42, 349–352. [Google Scholar] [CrossRef]
- Nacef, S.; Jannet, H.B.; Abreu, P.; Mighri, Z. Phenolic Constituents of Convolvulus dorycnium L. Flowers. Phytochem. Lett. 2010, 3, 66–69. [Google Scholar] [CrossRef]
- Zhang, L.; Tu, Z.; Yuan, T.; Wang, H.; Xie, X.; Fu, Z. Antioxidants and α-Glucosidase Inhibitors from Ipomoea batatas Leaves Identified by Bioassay-Guided Approach and Structure-Activity Relationships. Food Chem. 2016, 208, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ali, A.; Ali, J.; Sahni, J.K.; Baboota, S. Rutin: Therapeutic Potential and Recent Advances in Drug Delivery. Expert Opin. Investig. Drugs 2013, 22, 1063–1079. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Ku, S.-K.; Bae, J.-S. Barrier Protective Effects of Rutin in LPS-Induced Inflammation In Vitro and In Vivo. Food Chem. Toxicol. 2012, 50, 3048–3055. [Google Scholar] [CrossRef] [PubMed]
- Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A.A.; Khan, G.J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; FangFang, X.; Modarresi-Ghazani, F.; et al. Chlorogenic Acid (CGA): A Pharmacological Review and Call for Further Research. Biomed. Pharmacother. 2018, 97, 67–74. [Google Scholar] [CrossRef]
- Hwang, S.J.; Kim, Y.-W.; Park, Y.; Lee, H.-J.; Kim, K.-W. Anti-Inflammatory Effects of Chlorogenic Acid in Lipopolysaccharide-Stimulated RAW 264.7 Cells. Inflamm. Res. 2014, 63, 81–90. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, L.; Wang, M.-H. N-Trans-Feruloyltyramine Inhibits LPS-Induced NO and PGE2 Production in RAW 264.7 Macrophages: Involvement of AP-1 and MAP Kinase Signalling Pathways. Chem. Biol. Interact. 2015, 235, 56–62. [Google Scholar] [CrossRef]
- Le, T.B.; Beaufay, C.; Nghiem, D.T.; Mingeot-Leclercq, M.-P.; Quetin-Leclercq, J. In Vitro Anti-Leishmanial Activity of Essential Oils Extracted from Vietnamese Plants. Molecules 2017, 22, 1071. [Google Scholar] [CrossRef] [Green Version]
- Masquelier, J.; Alhouayek, M.; Terrasi, R.; Bottemanne, P.; Paquot, A.; Muccioli, G.G. Lysophosphatidylinositols in Inflammation and Macrophage Activation: Altered Levels and Anti-Inflammatory Effects. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1458–1468. [Google Scholar] [CrossRef]
- Alhouayek, M.; Bottemanne, P.; Makriyannis, A.; Muccioli, G.G. N-Acylethanolamine-Hydrolyzing Acid Amidase and Fatty Acid Amide Hydrolase Inhibition Differentially Affect N-Acylethanolamine Levels and Macrophage Activation. Biochim. Biophys Acta. Mol. Cell Biol. Lipids 2017, 1862, 474–484. [Google Scholar] [CrossRef]
- Alhouayek, M.; Masquelier, J.; Cani, P.D.; Lambert, D.M.; Muccioli, G.G. Implication of the Anti-Inflammatory Bioactive Lipid Prostaglandin D2-Glycerol Ester in the Control of Macrophage Activation and Inflammation by ABHD6. Proc. Natl. Acad. Sci. USA 2013, 110, 17558–17563. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]





| Code | tR [min] | λmax | m/z | MS Major Ion(s) | MS/MS Fragments [m/z] | Molecular Formula | Δ ppm | Δ mDa | Putative Identification | Isolated Previously a | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.20 | n.d. | 387.1147 | [M + HCOO−]− | 179.0563 b | C12H22O11 | 0.04 | 0.01 | Sucrose | ||
| 341.1084 | [M − H]− | ||||||||||
| 729.2287 | [2M + HCOO−]− | ||||||||||
| 683.2222 | [2M − H]− | ||||||||||
| 2 | 3.28 | n.d. | 151.0613 | [M − H]− | 71.0142; 101.0247; 133.0509 b | C5H12O5 | 4.31 | 0.65 | Xylitol # | ||
| 197.0664 | [M + HCOO−]− | ||||||||||
| 3 | 3.40 | n.d. | 181.0722 | [M + HCOO−]− | 135.0669 c | C5H12O4 | −2.01 | −0.27 | 2-methyl-1,2,3,4-butanetetrol # | ||
| 135.0668 | [M − H]− | ||||||||||
| 4 | 3.39 | 270 | 191.0567 | [M − H]− | 85.0299; 127.0405; 173.0458; 93.0350 b | C7H12O6 | 5.95 | 1.14 | Quinic acid # | C. althaeoides | [31] |
| 383.1199 | [2M − H]− | ||||||||||
| 5 | 3.59 | n.d. | 239.0775 | [M + HCOO−]− | 133.0509; 59.0141 b | C7H14O6 | 5.93 | 1.09 | O-methyl-inositol isomer I # | ||
| 193.0723 | [M − H]− | ||||||||||
| 6 | 3.96 | n.d. | 239.0772 | [M + HCOO]− | 193.0718; 133.0508; 59.0141 b | C7H14O6 | 2.12 | 0.51 | O-methyl-inositol isomer II # | ||
| 7 | 4.90 | n.d. | 239.0772 | [M + HCOO−]− | 133.0508; 59.0141 b | C7H14O6 | 3.04 | 0.59 | O-methyl-inositol isomer III # | ||
| 193.0718 | [M − H]− | ||||||||||
| 712.5355 | [M − H]− | ||||||||||
| 8 | 4.98 | n.d. | 117.0198 | [M − H]− | 73.0297; 99.0091 b | C4H6O4 | 8.69 | 1.02 | Succinic acid # | ||
| 9 | 5.06 | n.d. | 281.0881 | [M − H]− | 235.0820; 263.0955 b | C10H18O9 | 3.00 | 0.84 | Xylobiose # | ||
| 10 | 5.68 | n.d. | 341.0889 | [M − H]− | 179.0348; 167.0349; 135.0451 b | C15H18O9 | 4.82 | 1.64 | O-glucosyl-caffeic acid isomer # | ||
| 11 | 6.02 | n.d. | 451.2199 | [M + HCOO−]− | 167.1076; 179.0560; 243.1596 b | C19H34O9 | −3.60 | −1.46 | Magastigmane glycoside derivative I # | ||
| 405.2110 | [M − H]− | ||||||||||
| 12 | 6.18 | 289; 325 | 433.2091 | [M + HCOO−]− | 387.2012; 179.0559; 161.0454 c | C19H32O8 | 3.99 | 1.73 | Magastigmane glycoside derivative II # | ||
| 13 | 6.21 | n.d. | 353.0881 | [M − H]− | 191.0559; 179.0350; 173.0455; 135.0452 b | C16H18O9 | 2.39 | 0.84 | Chlorogenic acid * | C. arvensis; C. dorycnium | [32,33] |
| 14 | 7.30 | 315 | 369.0820 | [M + HCOO−]− | 323.0769; 161.0243 b | C15H16O8 | 0.64 | 0.21 | Skimmin # | Pharbitis nil | [34] |
| 323.0769 | [M − H]− | ||||||||||
| 15 | 7.97 | n.d. | 395.1930 | [M + HCOO−]− | 187.1341; 161.0457; 179.0563 b | C16H30O8 | 5.03 | 1.76 | Monoterpenoid glycoside I # | ||
| 349.1880 | [M − H]− | ||||||||||
| 16 | 8.66 | 245; 324 | 353.0881 | [M − H]− | 191.0560; 179.0350 b | C16H18O9 | 2.39 | 0.84 | O-caffeoylquinic acid # | Ipomoea batatas | [35] |
| 707.1868 | [2M − H]− | ||||||||||
| 17 | 9.06 | 288 | 433.2091 | [M + HCOO−]− | 161.0455 b | C19H32O8 | −2.88 | −1.12 | Magastigmane glycoside derivative III # | ||
| 387.2014 | [M − H]− | ||||||||||
| 18 | 10.20 | 287 | 387.1870 | [M − H]− | 207.1022; 163.1128; 369.1544 b | C15H32O11 | 0.94 | 0.36 | n.i. | ||
| 19 | 10.51 | n.d. | 431.1928 | [M + HCOO−]− | 223.1337; 205.1233; 153.0922; 161.0457 b | C19H30O8 | 1.97 | 0.76 | Roseoside # | Ipomoea purpurea | [36] |
| 385.1853 | [M − H]− | ||||||||||
| 20 | 11.36 | 322 | 297.0986 | [M − H]− | 179.0352; 135.0453 b | C14H18O7 | 3.95 | 1.17 | n.i. | ||
| 21 | 12.13 | 265 | 441.1969 | [M + HCOO−]− | n.s. | C17H32O10 | −4.61 | −1.82 | n.i. | ||
| 395.1899 | [M − H]− | ||||||||||
| 22 | 12.28 | n.d. | 281.1397 | [M − H]− | 237.1488; 171.1180; 123.0817; 201.1284 b | C15H22O5 | 2.85 | 0.80 | n.i. | ||
| 23 | 12.97 | 258 | 583.2027 | [M + HCOO−]− | n.s. | C26H34O12 | −1.49 | −0.80 | n.i. | ||
| 537.1964 | [M − H]− | ||||||||||
| 1187.3107 | [2M − H]− | ||||||||||
| 1613.8181 | [M − H]− | ||||||||||
| 24 | 13.26 | 242; 298; 324 | 179.0353 | [M − H]− | 135.0453 b | C9H8O4 | 4.84 | 0.87 | Caffeic acid # | C. trabutianus | [37] |
| 25 | 14.25 | 255; 331 | 741.1902 | [M − H]− | 300.0269; 609.1447; 301.0347; 591.1343; 271.0242 b | C32H38O20 | 3.21 | 2.38 | Quercetin-O-pentosyl-hexosyl-hexoside # | ||
| 26 | 14.33 | 294; 326 | 367.1024 | [M − H]− | 191.0559; 173.0455; 193.0499 b | C17H20O9 | −1.38 | −0.51 | Feruloyl quinic acid # | ||
| 27 | 14.78 | n.d. | 225.1138 | [M − H]− | 181.1234; 165.0921; 147.0816; 135.0816 b | C12H18O4 | 4.96 | 1.12 | Tuberonic acid # | Dichondra repens | [38] |
| 28 | 16.09 | n.d. | 725.1935 | [M − H]− | 593.1494; 575.1391; 284.0317; 285.0394; 327.0500 b | C32H38O19 | 0.82 | 0.60 | Kaempferol-O-pentosyl-hexosyl-hexoside # | ||
| 29 | 16.54 | 254; 347 | 609.1460 | [M − H]− | 301.0349; 300.0276; 271.0248; 343.0454 b | C27H30O16 | 0.72 | 0.44 | Rutin * | C. arvensis; Cressa cretica | [15,39] |
| 1219.3008 | [2M − H]− | ||||||||||
| 30 | 16.78 | n.d. | 621.3154 | [M + HCOO−]− | 557.2985 b | C28H48O12 | 3.91 | 2.25 | n.i. | ||
| 575.3090 | [M − H]− | ||||||||||
| 31 | 17.24 | n.d. | 665.3405 | [M + HCOO−]− | 619.3352; 601.3245 c | C31H54O15 | 3.61 | 2.23 | n.i. | ||
| 32 | 17.84 | n.d. | 709.3664 | [M + HCOO−]− | 645.3510; 663.3615 c | C32H56O14 | 2.45 | 1.74 | n.i. | ||
| 33 | 18.00 | 240; 301; 325 | 593.1503 | [M − H]− | 285.0398; 284.0325; 327.0507; 255.0297 b | C27H30O15 | −0.58 | −0.35 | Kaempferol-O-hexosyl-pentoside # | ||
| 34 | 18.36 | 309 | 753.3912 | [M + HCOO−]− | 689.3768; 707.3875 c | C34H60O15 | 0.43 | 0.32 | n.i. | ||
| 35 | 18.73 | 238; 265; 342 | 593.1503 | [M − H]− | 285.0369; 327.0500 b | C27H30O15 | −0.58 | −0.35 | Kaempferol-3-O-rutinoside *# | C. dorycnium | [40] |
| 1187.3107 | [2M − H]− | ||||||||||
| 36 | 19.20 | 297; 342 | 607.1292 | [M − H]− | 463.0877; 505.0982; 545.1293; 301.0352 b | C27H28O16 | −1.17 | −0.71 | Quercetin-O-[-O-(hydroxy-3-methylglutaryl)-hexoside] # | ||
| 37 | 19.56 | 301; 325 | 447.0950 | [M − H]− | 284.0324; 285.0399; 327.0507; 255.0297 b | C21H20O11 | 5.06 | 2.26 | Kaempferol-3-O-glucoside *# | C. trabutianus | [37] |
| 38 | 19.69 | 238; 325 | 515.1215 | [M − H]− | 353.0858; 173.0454; 335.0765; 179.0347 b | C25H24O12 | 4.95 | 2.55 | 3,4-di-O-caffeoylquinic acid # | C. trabutianus | [37] |
| 1031.2482 | [2M − H]− | ||||||||||
| 39 | 20.06 | n.d. | 511.2174 | [M + HCOO−]− | 161.0245; 179.0350 c | C24H34O9 | −1.05 | −0.54 | n.i. | ||
| 929.4974 | [2M − H]− | ||||||||||
| 40 | 20.52 | n.d. | 973.5257 | [M − H]− | 909.508 b | C46H78N4O18 | 2.48 | 2.41 | n.i. | ||
| 41 | 20.96 | 245; 295; 327 | 515.1211 | [M − H]− | 353.0868; 191.056; 179.0349 b | C25H24O12 | 4.17 | 2.15 | 3,5-di-O-caffeoylquinic acid *# | C. trabutianus | [37] |
| 561.1236 | [M + HCOO−]− | ||||||||||
| 1031.2471 | [2M − H]− | ||||||||||
| 42 | 21.67 | n.d. | 417.0831 | [M − H]− | 284.0323; 285.0397; 327.0506; 255.0295 b | C20H18O10 | 2.23 | 1.80 | Kaempferol-O-pentoside # | ||
| 43 | 21.86 | 245; 293; 324 | 515.1214 | [M − H]− | 353.0866; 299.056; 173.0457; 203.0350; 191.0560 b | C25H24O12 | 2.37 | 0.93 | 4,5-di-O-caffeoylquinic acid # | ||
| 1031.2463 | [2M − H]− | ||||||||||
| 44 | 22.49 | n.d. | 187.0981 | [M − H]− | 125.0975; 97.0663 b | C9H16O4 | 5.70 | 1.07 | Azelaic acid # | ||
| 45 | 22.93 | n.d. | 625.2126 | [M + HCOO−]− | n.s. | C28H36O13 | −0.98 | −0.57 | n.i. | ||
| 579.2072 | [M − H]− | ||||||||||
| 46 | 24.91 | n.d. | 529.1365 | [M − H]− | 353.0868; 367.1023; 191.0561 b | C26H26O12 | 3.59 | 1.90 | Caffeoyl-feruloyl quinic acid isomer I # | ||
| 47 | 25.41 | n.d. | 529.1372 | [M − H]− | 367.1016; 173.0454; 193.0506; 179.0349 b | C26H26O12 | 4.91 | 2.60 | Caffeoyl-feruloyl quinic acid isomer II # | ||
| 48 | 26.09 | 292; 305 | 282.1146 | [M − H]− | 119.0506; 145.0296; 162.0563 b | C17H17NO3 | 5.61 | 1.58 | N-trans-p-coumaroyltyramine *# | Ipomoea batatas | [41] |
| 328.1194 | [M + HCOO−]− | ||||||||||
| 49 | 26.67 | 238; 293; 318 | 312.1243 | [M − H]− | 178.0512; 297.1007; 135.0454 b | C18H19NO4 | 2.30 | 0.72 | N-trans-feruloyltyramine *# | Ipomoea batatas | [41] |
| 358.1294 | [M + HCOO−]− | ||||||||||
| 50 | 28.04 | n.d. | 607.2026 | [M + HCOO−]− | 561.1960 c | C28H34O12 | −0.13 | −0.08 | n.i. | ||
| 51 | 29.36 | n.d. | 327.2182 | [M − H]− | 229.1438; 291.1964, 211.1336; 171.1029 b | C18H32O5 | 3.21 | 1.05 | Trihydroxy-10,15-octadecadienoic acid derivative I # | ||
| 52 | 30.56 | n.d. | 327.2178 | [M − H]− | 229.1437; 291.1961; 171.1027; 211.1336; 309.2067 b | C18H32O5 | 1.99 | 0.65 | Trihydroxy-10,15-octadecadienoic acid derivative II # | ||
| 53 | 31.36 | n.d. | 329.2329 | [M − H]− | 329.2333; 229.1440; 211.1337; 171.1030; 311.2228; 293.2122 b | C18H34O5 | 0.31 | 0.10 | Trihydroxy-10-octadecenoic acid # | ||
| 54 | 31.90 | n.d. | 883.4227 | [M + HCOO−]− | 561.6332; 533.2184 b | C39H66O19 | 2.62 | 2.20 | O-(Hexosyl-hexosyl-hexosyl)-octadecatrienoyl-glycerol # | ||
| 837.4142 | [M − H]− | ||||||||||
| 55 | 32.54 | n.d. | 721.3635 | [M + HCOO−]− | 397.1340 b | C33H56O14 | −0.86 | −0.58 | O-(Hexosyl-hexosyl)-O-linolenoyl-glycerol # | ||
| 675.3586 | [M − H]− | ||||||||||
| 712.5355 | [M − H]− | ||||||||||
| 56 | 33.05 | n.d. | 647.3270 | [M + HCOO−]− | n.s. | C30H50O12 | −1.67 | −1.00 | n.i. | ||
| 601.3214 | [M − H]− | ||||||||||
| 57 | 33.53 | 218 | 559.3091 | [M + HCOO−]− | 277.2170; 253.0926 c | C27H46O9 | −4.89 | −2.74 | O-Hexosyl-O-linolenoyl-glycerol # | ||
| 58 | 34.56 | 215 | 699.3825 | [M + HCOO]− | 397.1340 c | C31H58O14 | 3.13 | 2.19 | O-(Hexosyl-hexosyl)-O-palmitoyl-glycerol # | ||
| 59 | 35.20 | n.d. | 1659.8471 | [M + HCOO−]− | n.s. | C98H118O20 | 2.65 | 4.28 | n.i. | ||
| 1613.8181 | [M − H]− | ||||||||||
| 60 | 35.87 | 218 | 819.5250 | [M + HCOO−]− | 773.5198; 277.2170; 513.3065 b | C45H74O10 | −0.74 | −0.57 | O-Hexosyl-di-O-linolenoyl-glycerol # | ||
| 773.5198 | [M − H]− | ||||||||||
| 61 | 38.25 | 409 | 997.5783 | [M + HCOO−]− | n.s. | C51H84O16 | 4.40 | 4.19 | n.i. | ||
| 951.5723 | [M − H]− | ||||||||||
| 62 | 38.70 | 218 | 765.5196 | [M − H]− | 505.3003; 255.2320; 277.2163; 527.2846; 747.5024 b | C43H74O11 | 5.63 | 4.31 | n.i. | ||
| 63 | 40.11 | 409 | 591.2607 | [M − H]− | 559.2329; 515.2441 b | C34H40O9 | 2.19 | 1.29 | n.i. | ||
| 64 | 40.69 | n.d. | 1835.8597 | [M − H]− | n.s. | C116H124O20 | −0.58 | −1.07 | n.i. | ||
| 65 | 41.44 | n.d. | 758.5413 | [M + HCOO−]− | n.s. | C41H71N5O5 | −3.08 | −2.20 | n.i. | ||
| 712.5355 | [M − H]− |
| ID | Compound | Regression Equation | R2 | LOD (µg/mL) | LOQ (µg/mL) | ARE | ARM | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Conc. in Crude Extract (mg/g Dry Extract) | Concentration in 50 µg of Extract/mL | Conc. in Crude Extract (mg/g Dry Extract) | Concentration in 50 µg of Extract/mL | ||||||||
| (µg/mL) | (µM) | (µg/mL) | (µM) | ||||||||
| 13 | Chlorogenic acid | y = 40320x + 99146 | 0.9996 | 4.37 | 13.24 | 1.51 ± 0.04 | 0.08 ± 0.00 | 0.21 ± 0.01 | 29.86 ± 0.17 | 1.49 ± 0.01 | 4.22 ± 0.02 |
| 29 | Rutin | y = 26787x + 147669 | 0.9997 | 5.11 | 15.48 | 0.78 ± 0.19 | 0.04 ± 0.01 | 0.06 ± 0.02 | 27.71 ± 0.19 | 1.39 ± 0.01 | 2.27 ± 0.02 |
| 35 | Kaempferol-3-O-rutinoside | y = 36219x + 52756 | 0.9996 | 4.16 | 12.60 | 2.18 ± 0.02 | 0.11 ± 0.00 | 0.18 ± 0.00 | 5.23 ± 0.19 | 0.26 ± 0.01 | 0.44 ± 0.02 |
| 41 | 3,5-di-O-caffeoylquinic acid | y = 59073x + 308772 | 0.9971 | 8.68 | 26.32 | 5.82 ± 0.32 | 0.29 ± 0.02 | 0.56 ± 0.03 | 38.16 ± 0.13 | 1.91 ± 0.01 | 3.70 ± 0.01 |
| 48 | N-trans-p-coumaroyltyramine | y = 88859x + 21246 | 0.9997 | 1.81 | 5.47 | 5.90 ± 0.27 | 0.30 ± 0.01 | 1.04 ± 0.05 | 1.72 ± 0.13 | 0.09 ± 0.01 | 0.30 ± 0.02 |
| ID | Compound | ARE | ARM | ||||
|---|---|---|---|---|---|---|---|
| Conc. in Crude Extract (mg/g Dry Extract) | Concentration in 50 µg of Extract/mL | Conc. in Crude Extract (mg/g Dry Extract) | Concentration in 50 µg of Extract/mL | ||||
| (µg/mL) | (µM) | (µg/mL) | (µM) | ||||
| 38 | 3,4-di-O-caffeoylquinic acid | 1.48 ± 0.04 a | 0.07 ± 0.00 a | 0.14 ± 0.00 a | 14.08 ± 0.81a | 0.704 ± 0.04 a | 1.36 ± 0.08 a |
| 43 | 4,5-di-O-caffeoylquinic acid | 1.97 ± 0.03 a | 0.10 ± 0.00 a | 0.19 ± 0.00 a | 21.34 ± 0.06 a | 1.067 ± 0.00 a | 2.07 ± 0.01 a |
| 49 | N-trans-feruloyltyramine | 6.61 ± 0.40 b | 0.33 ± 0.02 b | 1.06 ± 0.06 b | 1.37 ± 0.30 b | 0.068 ± 0.02 b | 0.22 ± 0.05 b |
| Gene/Product | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| Il6/IL-6 | ACAAGTCGGAGGCTTAATTACACAT | TTGCCATTGCACAACTCTTTTC |
| Tnf/TNF-α | CTACTGAACTTCGGGGTGATC | TGAGTGTGAGGGTCTGGGC |
| Ccl2/MCP-1 | GTCCCAAAGAAGCTGTAGTTTTTG | ATGTATGTCTGGACCCATTCC |
| Rpl19/RPL19 | TGACCTGGATGAGAAGGATGAG | CTGTGATACATATGGCGGTCAATC |
| Ptgs2/COX-2 | TGACCCCCAAGGCTCAAATAT | TGAACCCAGGTCCTCGCTTA |
| Nos2/iNOS | AGGTACTCAGCGTGCTCCAC | GCACCGAAGATATCTTCATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Khaliq, H.; Ortiz, S.; Alhouayek, M.; Muccioli, G.G.; Quetin-Leclercq, J. Dereplication and Quantification of Major Compounds of Convolvulus arvensis L. Extracts and Assessment of Their Effect on LPS-Activated J774 Macrophages. Molecules 2022, 27, 963. https://doi.org/10.3390/molecules27030963
Abdul Khaliq H, Ortiz S, Alhouayek M, Muccioli GG, Quetin-Leclercq J. Dereplication and Quantification of Major Compounds of Convolvulus arvensis L. Extracts and Assessment of Their Effect on LPS-Activated J774 Macrophages. Molecules. 2022; 27(3):963. https://doi.org/10.3390/molecules27030963
Chicago/Turabian StyleAbdul Khaliq, Hafiz, Sergio Ortiz, Mireille Alhouayek, Giulio G. Muccioli, and Joëlle Quetin-Leclercq. 2022. "Dereplication and Quantification of Major Compounds of Convolvulus arvensis L. Extracts and Assessment of Their Effect on LPS-Activated J774 Macrophages" Molecules 27, no. 3: 963. https://doi.org/10.3390/molecules27030963