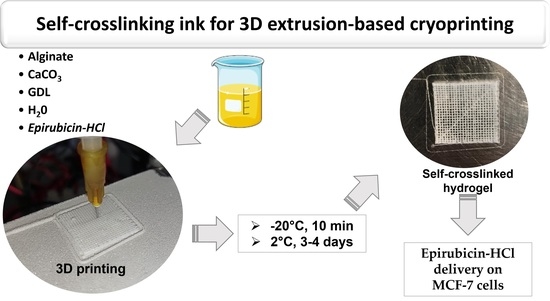
Alginate Self-Crosslinking Ink for 3D Extrusion-Based Cryoprinting and Application for Epirubicin-HCl Delivery on MCF-7 Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the Self-Crosslinking Formulation for 3D Printing
2.2. ATR FT-IR Scaffold Characterization
2.3. Swelling Behavior and Drug Release from 3D-Printed Hydrogel
2.4. Cytotoxic Activity towards MCF-7 Breast-Cancer Cells
2.5. Proteomic Analysis
3. Materials and Methods
3.1. Reagents
3.2. Ink Preparation for 3D Printing
3.3. Scaffold Design
3.4. 3D Printer and Scaffold Manufacturing
3.5. Swelling Test
3.6. Attenuated Total Reflection (ATR) Fourier Transform Infrared (FT-IR) Spectroscopy
3.7. Evaluation of the Epirubicin-HCl Loading
3.8. In Vitro Drug Release Studies
3.9. MCF7 Cells Culture
3.10. Cytotoxic Activity towards MCF-7 Breast-Cancer Cells
3.11. Proteomic Analysis
3.12. Bioinformatic Analysis and Mass Spectrometry Data Processing
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, J.; Zhang, Y.; Aghda, N.H.; Pillai, A.R.; Thakkar, R.; Nokhodchi, A.; Maniruzzaman, M. Emerging 3D Printing Technologies for Drug Delivery Devices: Current Status and Future Perspective. Adv. Drug Deliv. Rev. 2021, 174, 294–316. [Google Scholar] [CrossRef]
- Jiang, Z.; Diggle, B.; Tan, M.L.; Viktorova, J.; Bennett, C.W.; Connal, L.A. Extrusion 3D Printing of Polymeric Materials with Advanced Properties. Adv. Sci. 2020, 7, 2001379. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Nair, A.B.; Patel, V.; Shah, J. 3D Printing Technologies: Recent Development and Emerging Applications in Various Drug Delivery Systems. AAPS PharmSciTech 2020, 21, 220. [Google Scholar] [CrossRef]
- Kotta, S.; Nair, A.; Alsabeelah, N. 3D Printing Technology in Drug Delivery: Recent Progress and Application. Curr. Pharm. Des. 2018, 24, 5039–5048. [Google Scholar] [CrossRef]
- Li, J.; Wu, C.; Chu, P.K.; Gelinsky, M. 3D Printing of Hydrogels: Rational Design Strategies and Emerging Biomedical Applications. Mater. Sci. Eng. R Rep. 2020, 140, 100543. [Google Scholar] [CrossRef]
- Hoffman, A.S. Hydrogels for Biomedical Applications. Adv. Drug Deliv. Rev. 2012, 64, 18–23. [Google Scholar] [CrossRef]
- Wei, X.; Liu, C.; Wang, Z.; Luo, Y. 3D Printed Core-Shell Hydrogel Fiber Scaffolds with NIR-Triggered Drug Release for Localized Therapy of Breast Cancer. Int. J. Pharm. 2020, 580, 119219. [Google Scholar] [CrossRef]
- Bergonzi, C.; Bianchera, A.; Remaggi, G.; Ossiprandi, M.C.; Zimetti, F.; Marchi, C.; Bernini, F.; Bettini, R.; Elviri, L. Biocompatible 3D Printed Chitosan-Based Scaffolds Containing α-Tocopherol Showing Antioxidant and Antimicrobial Activity. Appl. Sci. 2021, 11, 7253. [Google Scholar] [CrossRef]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The Bioink: A Comprehensive Review on Bioprintable Materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and Biomedical Applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [Green Version]
- Grant, G.T.; Mon, E.R.; Rees, S.D.A. Biological Interactions Between Polysacchartdes and Divalent Cations: The Egg-Box Model. FEBS Lett. 1973, 32, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Lu, W.; Mata, A.; Nishinari, K.; Fang, Y. Egg-Box Model-Based Gelation of Alginate and Pectin: A Review. Carbohydr. Polym. 2020, 242, 116389. [Google Scholar] [CrossRef] [PubMed]
- Aguero, L.; Alpdagtas, S.; Ilhan, E.; Zaldivar-Silva, D.; Gunduz, O. Functional Role of Crosslinking in Alginate Scaffold for Drug Delivery and Tissue Engineering: A Review. Eur. Polym. J. 2021, 160, 110807. [Google Scholar] [CrossRef]
- Gao, Q.; He, Y.; Fu, J.; Liu, A.; Ma, L. Coaxial Nozzle-Assisted 3D Bioprinting with Built-in Microchannels for Nutrients Delivery. Biomaterials 2015, 61, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.K.; Ma, P.X. Maintaining Dimensions and Mechanical Properties of Ionically Crosslinked Alginate Hydrogel Scaffoldsin Vitro. J. Biomed. Mater. Res. A 2008, 84, 899–907. [Google Scholar] [CrossRef] [Green Version]
- Catanzano, O.; D’Esposito, V.; Acierno, S.; Ambrosio, M.R.; De Caro, C.; Avagliano, C.; Russo, P.; Russo, R.; Miro, A.; Ungaro, F.; et al. Alginate–Hyaluronan Composite Hydrogels Accelerate Wound Healing Process. Carbohydr. Polym. 2015, 131, 407–414. [Google Scholar] [CrossRef] [PubMed]
- GhavamiNejad, A.; Ashammakhi, N.; Wu, X.Y.; Khademhosseini, A. Crosslinking Strategies for 3D Bioprinting of Polymeric Hydrogels. Small 2020, 16, 2002931. [Google Scholar] [CrossRef] [PubMed]
- Catanzano, O.; D′Esposito, V.; Formisano, P.; Boateng, J.S.; Quaglia, F. Composite Alginate-Hyaluronan Sponges for the Delivery of Tranexamic Acid in Postextractive Alveolar Wounds. J. Pharm. Sci. 2018, 107, 654–661. [Google Scholar] [CrossRef]
- Hazur, J.; Detsch, R.; Karakaya, E.; Kaschta, J.; Teßmar, J.; Schneidereit, D.; Friedrich, O.; Schubert, D.W.; Boccaccini, A.R. Improving Alginate Printability for Biofabrication: Establishment of a Universal and Homogeneous Pre-Crosslinking Technique. Biofabrication 2020, 12, 045004. [Google Scholar] [CrossRef]
- Sun, W.-L.; Wang, L.; Luo, J.; Zhu, H.-W.; Cai, Z.-W. Ambra1 Modulates the Sensitivity of Breast Cancer Cells to Epirubicin by Regulating Autophagy via ATG12. Cancer Sci. 2018, 109, 3129–3138. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Kumari, R.M.; Gupta, N.; Syed, A.; Bahkali, A.H.; Nimesh, S. Poly-(Lactic-Co-Glycolic) Acid Nanoparticles for Synergistic Delivery of Epirubicin and Paclitaxel to Human Lung Cancer Cells. Molecules 2020, 25, 4243. [Google Scholar] [CrossRef]
- Conte, P.F.; Gennari, A.; Landucci, E.; Orlandini, C. Role of Epirubicin in Advanced Breast Cancer. Clin. Breast Cancer 2000, 1, S46–S51. [Google Scholar] [CrossRef]
- Perveen, K.; Masood, F.; Hameed, A. Preparation, Characterization and Evaluation of Antibacterial Properties of Epirubicin Loaded PHB and PHBV Nanoparticles. Int. J. Biol. Macromol. 2020, 144, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Ansari, L.; Derakhshi, M.; Bagheri, E.; Shahtahmassebi, N.; Malaekeh-Nikouei, B. Folate Conjugation Improved Uptake and Targeting of Porous Hydroxyapatite Nanoparticles Containing Epirubicin to Cancer Cells. Pharm. Dev. Technol. 2020, 25, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-W.; Wu, Y.-T.; Lin, K.-J.; Yu, T.-J.; Kuo, Y.-L.; Chang, L.-C. A Hydrogel-Based Epirubicin Delivery System for Intravesical Chemotherapy. Molecules 2016, 21, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, R.; Khatri, N.; Baradia, D.; Vhora, I.; Misra, A. Surface-Modified Epirubicin-HCl Liposomes and Its in Vitro Assessment in Breast Cancer Cell-Line: MCF-7. Drug Deliv. 2016, 23, 1152–1162. [Google Scholar] [CrossRef] [Green Version]
- Matai, I.; Gopinath, P. Chemically Cross-Linked Hybrid Nanogels of Alginate and PAMAM Dendrimers as Efficient Anticancer Drug Delivery Vehicles. ACS Biomater. Sci. Eng. 2016, 2, 213–223. [Google Scholar] [CrossRef]
- Bahreyni, A.; Alibolandi, M.; Ramezani, M.; Sarafan Sadeghi, A.; Abnous, K.; Taghdisi, S.M. A Novel MUC1 Aptamer-Modified PLGA-Epirubicin-PβAE-Antimir-21 Nanocomplex Platform for Targeted Co-Delivery of Anticancer Agents in Vitro and in Vivo. Colloids Surf. B Biointerfaces 2019, 175, 231–238. [Google Scholar] [CrossRef]
- Zanotti, I.; Marando, S.; Remaggi, G.; Bergonzi, C.; Bernini, F.; Bettini, R.; Elviri, L. The Adaptation of Lipid Profile of Human Fibroblasts to Alginate 2D Films and 3D Printed Scaffolds. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2021, 1865, 129734. [Google Scholar] [CrossRef]
- Bergonzi, C.; Remaggi, G.; Graiff, C.; Bergamonti, L.; Potenza, M.; Ossiprandi, M.C.; Zanotti, I.; Bernini, F.; Bettini, R.; Elviri, L. Three-Dimensional (3D) Printed Silver Nanoparticles/Alginate/Nanocrystalline Cellulose Hydrogels: Study of the Antimicrobial and Cytotoxicity Efficacy. Nanomaterials 2020, 10, 844. [Google Scholar] [CrossRef]
- Elviri, L.; Foresti, R.; Bergonzi, C.; Zimetti, F.; Marchi, C.; Bianchera, A.; Bernini, F.; Silvestri, M.; Bettini, R. Highly Defined 3D Printed Chitosan Scaffolds Featuring Improved Cell Growth. Biomed. Mater. 2017, 12, 045009. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, G.; Keen, I.; Drew, B.; Chandler-Temple, A.; Rintoul, L.; Fredericks, P.; Grøndahl, L. Interactions between Alginate and Chitosan Biopolymers Characterized Using FTIR and XPS. Biomacromolecules 2007, 8, 2533–2541. [Google Scholar] [CrossRef]
- Fu, Y.; Kao, W.J. Drug Release Kinetics and Transport Mechanisms of Non-Degradable and Degradable Polymeric Delivery Systems. Expert Opin. Drug Deliv. 2010, 7, 429–444. [Google Scholar] [CrossRef]
- Li, J.; Mooney, D.J. Designing Hydrogels for Controlled Drug Delivery. Nat. Rev. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, D.; Saini, B.; Bansal, G. Identification of Four New Degradation Products of Epirubicin Through Forced Degradation, LC–UV, MS n and LC–MS–TOF Studies. J. Chromatogr. Sci. 2015, 53, 1737–1748. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Tang, Q.; Fang, D.; Xiong, G.; Singla, N.; He, Q.; Zhang, L.; Liu, P.; Fan, Y.; Hao, H.; et al. Comparisons of Prognosis between Urothelial Carcinoma of the Upper Urinary Tract and Bladder with PT3-4 Cancer. Int. J. Clin. Exp. Med. 2016, 9, 18308–18315. [Google Scholar] [PubMed]
- ISO 10993-5:2009; Biological Evaluation of Medical Devices—Part 5: Tests for Citotoxicity, in Vitro Methods 2009. ISO: Geneva, Switzerland, 2009.
- Sun, W.-L.; Chen, J.; Wang, Y.-P.; Zheng, H. Autophagy Protects Breast Cancer Cells from Epirubicin-Induced Apoptosis and Facilitates Epirubicin-Resistance Development. Autophagy 2011, 7, 1035–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvajal, L.A.; Manfredi, J.J. Another Fork in the Road—Life or Death Decisions by the Tumour Suppressor P53. EMBO Rep. 2013, 14, 414–421. [Google Scholar] [CrossRef] [Green Version]
- An, H.-K.; Chung, K.M.; Park, H.; Hong, J.; Gim, J.-E.; Choi, H.; Lee, Y.W.; Choi, J.; Mun, J.Y.; Yu, S.-W. CASP9 (Caspase 9) Is Essential for Autophagosome Maturation through Regulation of Mitochondrial Homeostasis. Autophagy 2020, 16, 1598–1617. [Google Scholar] [CrossRef]
- Bandiera, A.; Catanzano, O.; Bertoncin, P.; Bergonzi, C.; Bettini, R.; Elviri, L. 3D-printed Scaffold Composites for the Stimuli-induced Local Delivery of Bioactive Adjuncts. Biotechnol. Appl. Biochem. 2021, 1–12. [Google Scholar] [CrossRef]






| (GDL) mM | Ink Working Time * (Min) | Number of Layers | Curing at 2 °C | Detachment from Plate | Self Cross-Linking |
|---|---|---|---|---|---|
| 180 | 8 | 2 | 1 | no | yes |
| 160 | 11 | 3 | 1 | no | yes |
| 2; 3; 4 | yes | ||||
| 5 | difficult | ||||
| 140 | 14 | 4 | 1 | no | no |
| 2 | no | yes | |||
| 3 | yes | yes | |||
| 4 | yes | yes | |||
| 5 | difficult | yes | |||
| 120 | 18 | 6 | 1 | no | no |
| 2 | no | ||||
| 3 | yes | ||||
| 4; 5 | yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remaggi, G.; Catanzano, O.; Quaglia, F.; Elviri, L. Alginate Self-Crosslinking Ink for 3D Extrusion-Based Cryoprinting and Application for Epirubicin-HCl Delivery on MCF-7 Cells. Molecules 2022, 27, 882. https://doi.org/10.3390/molecules27030882
Remaggi G, Catanzano O, Quaglia F, Elviri L. Alginate Self-Crosslinking Ink for 3D Extrusion-Based Cryoprinting and Application for Epirubicin-HCl Delivery on MCF-7 Cells. Molecules. 2022; 27(3):882. https://doi.org/10.3390/molecules27030882
Chicago/Turabian StyleRemaggi, Giulia, Ovidio Catanzano, Fabiana Quaglia, and Lisa Elviri. 2022. "Alginate Self-Crosslinking Ink for 3D Extrusion-Based Cryoprinting and Application for Epirubicin-HCl Delivery on MCF-7 Cells" Molecules 27, no. 3: 882. https://doi.org/10.3390/molecules27030882