Identification of Anti-Proliferative Compounds from Genista monspessulana Seeds through Covariate-Based Integration of Chemical Fingerprints and Bioactivity Datasets
Abstract
:1. Introduction
2. Results and Discussion
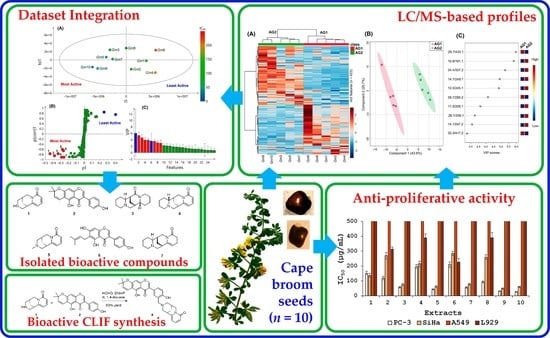
2.1. Anti-Proliferative Activity of G. monspessulana Seed-Derived Extracts
2.2. Characterization Based on LC-ESI-MS Data of G. monspessulana Seed-Derived Extracts
2.3. Detection of Anti-Proliferative Candidates from G. monspessulana Accessions through the Integration of Chemical Fingerprint and Bioactivity Datasets
2.4. Isolation and Identification of OPLS-Recognized Anti-Proliferative Candidates
3. Materials and Methods
3.1. Plant Material
3.2. Extract Preparation
3.3. In Vitro Cell Viability Assay
3.4. High-Performance Liquid Chromatography Coupled to Mass Spectrometry
3.5. LC-MS-Derived Fingerprint Processing
3.6. Multiple-Covariate Integration of Chemical Fingerprint and Bioactivity Datasets
3.7. Purification and Identification of Anti-Proliferatives 1–7 by Semipreparative HPLC
3.8. Synthesis of Cytisine-Linked Isoflavonoid 8
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Martin, E.; Dinc, M.; Duran, A.; Dogan, B.; Hakki, E.E. Cytogenetical analysis of 12 taxa of Genista L. (Fabaceae) from Turkey. Bangladesh J. Plant Taxon. 2009, 16, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Norverto, C.A.; González-Andrés, F.; Ortiz, J.M. Leaf and Stem Anatomy of Species of Cytisophyllum, Cytisus, Chamaecytisus, Genista, and Genista sect. Teline (Fabaceae: Genisteae) as an Aid for Taxonomy. Isr. J. Plant Sci. 1994, 42, 213–225. [Google Scholar] [CrossRef]
- Sheppard, A.W.; Henry, K. Genista monspessulana (L.) L. Johnson–Cape Broom. In Biological Control of Weeds in Australia; Julien, M., McFadyen, R., Cullen, R., Eds.; CSIRO Publishing: Melbourne, Victoria, Australia, 2012; pp. 267–273. ISBN 9780643104204. [Google Scholar]
- Gomez, P.; Bustamante, R.; Martín, J. Population structure of Teline monspessulana (L.) K. Koch in fragments of maulino forest in Central Chile. Gayana Bot. 2012, 69, 197–200. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, P. A success story: The Cape broom psyllid, Arytinnis hakani Loginova. Plant Prot. Q. 2013, 28, 81–82. [Google Scholar]
- Herrera, A.M.; Carruthers, R.I.; Mills, N.J. Introduced populations of Genista monspessulana (French broom) are more dense and produce a greater seed rain in California, USA, than native populations in the Mediterranean Basin of Europe. Biol. Invasions 2011, 13, 369–380. [Google Scholar] [CrossRef]
- Meza, A.; Rojas, P.; Cely-Veloza, W.; Guerrero-Perilla, C.; Coy-Barrera, E. Variation of isoflavone content and DPPH• scavenging capacity of phytohormone-treated seedlings after in vitro germination of cape broom (Genista monspessulana). S. Afr. J. Bot. 2020, 130, 64–74. [Google Scholar] [CrossRef]
- García, R.A.; Pauchard, A.; Cavieres, L.A.; Peña, E.; Rodríguez, M.F. Fire promotes Teline monspessulana (Fabaceae) invasion by increasing its germination. Rev. Chil. Hist. Nat. 2010, 83, 443–452. [Google Scholar]
- Cook, B.S.; Smith, L. Prerelease efficacy test of the psyllid, Arytinnis hakani, a prospective biological control agent of the invasive weed Genista monspessulana. Biocontrol Sci. Technol. 2014, 24, 641–651. [Google Scholar] [CrossRef]
- Geerts, S.; Botha, P.W.; Visser, V.; Richardson, D.M.; Wilson, J.R.U. Montpellier broom (Genista monspessulana) and Spanish broom (Spartium junceum) in South Africa: An assessment of invasiveness and options for management. S. Afr. J. Bot. 2013, 87, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Bitume, E.V.; Moran, P.J.; Sforza, R.F.H. Impact in quarantine of the galling weevil Lepidapion argentatum on shoot growth of French broom (Genista monspessulana), an invasive weed in the western U.S. Biocontrol Sci. Technol. 2019, 29, 615–625. [Google Scholar] [CrossRef]
- Kerdellant, E.; Thomann, T.; Sheppard, A.; Sforza, R.F.H. Host Specificity and Preliminary Impact of Lepidapion argentatum (Coleoptera, Brentidae), a Biocontrol Candidate for French Broom (Genista monspessulana, Fabaceae). Insects 2021, 12, 691. [Google Scholar] [CrossRef] [PubMed]
- Shah, I.; Shah, M.A.; Nawaz, M.A.; Pervez, S.; Noreen, N.; Vargas-de la Cruz, C.; Khan, F.; Blundell, R.; Briffa, J.; Azzopardi, J.; et al. Analysis of other phenolics (capsaicin, gingerol, and alkylresorcinols). In Recent Advances in Natural Products Analysis; Silva, A.S., Nabavi, S.F., Saeedi, M., Nabavi, S.M., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 6, pp. 255–271. ISBN 9780128164556. [Google Scholar]
- Ramos, L.; Apráez, J.; Cortes, K.; Apráez, J. Nutritional, antinutritional and phenological characterization of promising forage species for animal feeding in a cold tropical zone. Rev. Cienc. Agrícolas 2021, 38, 86–96. [Google Scholar] [CrossRef]
- Osorio-Castiblanco, D.F.; Peyre, G.; Saldarriaga, J.F. Physicochemical Analysis and Essential Oils Extraction of the Gorse (Ulex europaeus) and French Broom (Genista monspessulana), Two Highly Invasive Species in the Colombian Andes. Sustainability 2020, 12, 57. [Google Scholar] [CrossRef] [Green Version]
- Bisby, F. Plants and their constituents. In Phytochemical Dictionary of the Leguminosae, 1st ed.; Chapman and Hall: London, UK, 1994; Volume 1, pp. 313–330. ISBN 0412397706. [Google Scholar]
- Cheilari, A.; Vontzalidou, A.; Makropoulou, M.; Meligova, A.K.; Fokialakis, N.; Mitakou, S.; Alexis, M.N.; Aligiannis, N. Isoflavonoid Profiling and Estrogen-Like Activity of Four Genista Species from the Greek Flora. Molecules 2020, 25, 5507. [Google Scholar] [CrossRef]
- Pinto, D.C.G.A.; Simões, M.A.M.; Silva, A.M.S. Genista tridentata L.: A Rich Source of Flavonoids with Anti-inflammatory Activity. Medicines 2020, 7, 31. [Google Scholar] [CrossRef]
- Rauter, A.P.; Martins, A.; Lopes, R.; Ferreira, J.; Serralheiro, L.M.; Araújo, M.-E.; Borges, C.; Justino, J.; Silva, F.V.; Goulart, M.; et al. Bioactivity studies and chemical profile of the antidiabetic plant Genista tenera. J. Ethnopharmacol. 2009, 122, 384–393. [Google Scholar] [CrossRef] [Green Version]
- Barek, S.; Rahmoun, N.M.; Aissaoui, M.; El Haci, I.A.; Bensouici, C.; Choukchou-Braham, E.N. Phenolic Contents, Antioxidant, and Antibacterial Activities of the Algerian Genista saharae Solvent Extracts. J. Herbs. Spices Med. Plants 2020, 26, 1–13. [Google Scholar] [CrossRef]
- Bontempo, P.; Rigano, D.; Doto, A.; Formisano, C.; Conte, M.; Nebbioso, A.; Carafa, V.; Caserta, G.; Sica, V.; Molinari, A.M.; et al. Genista sessilifolia DC. extracts induce apoptosis across a range of cancer cell lines. Cell Prolif. 2013, 46, 183–192. [Google Scholar] [CrossRef]
- Sebaihi-Harzoun, S.; Atmani-Kilani, D.; Debbache-Benaida, N.; Nana, F.; Evain-Bana, E.; Kirsch, G.; Tabart, J.; Kevers, C.; Atmani, D. Phytochemical composition, antioxidant and anti-proliferative properties of Genista ferox Poirret. aerial parts. Eur. J. Integr. Med. 2018, 23, 6–13. [Google Scholar]
- Rigano, D.; Russo, A.; Formisano, C.; Cardile, V.; Senatore, F. Antiproliferative and Cytotoxic Effects on Malignant Melanoma Cells of Essential Oils from the Aerial Parts of Genista sessilifolia and G. tinctoria. Nat. Prod. Commun. 2010, 5, 1934578X1000500731. [Google Scholar] [CrossRef] [Green Version]
- Grafakou, M.-E.; Barda, C.; Tomou, E.-M.; Skaltsa, H. The genus Genista L.: A rich source of bioactive flavonoids. Phytochemistry 2021, 181, 112574. [Google Scholar] [CrossRef]
- Fritsche, S.; Steinhart, H. Occurrence of hormonally active compounds in food: A review. Eur. Food Res. Technol. 1999, 209, 153–179. [Google Scholar] [CrossRef]
- Boulanouar, B.; Abdelaziz, G.; Aazza, S.; Gago, C.; Miguel, M.G. Antioxidant activities of eight Algerian plant extracts and two essential oils. Ind. Crops Prod. 2013, 46, 85–96. [Google Scholar] [CrossRef]
- Łuczkiewicz, M.; Piotrowski, A. Two-stage system for micropropagation of several Genista plants producing large amounts of phytoestrogens. Z. Naturforsch. C 2005, 60, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Wink, M.; Montllor, C.B.; Bernays, E.A.; Witte, L. Uresiphita reversalis (Lepidoptera: Pyralidae): Carrier-Mediated Uptake and Sequestration of Quinolizidine Alkaloids Obtained from the Host Plant Teline monspessulana. Z. Naturforsch. C 1991, 46, 1080–1088. [Google Scholar] [CrossRef]
- Montllor, C.B.; Bernays, E.A.; Barbehenn, R. Importance of quinolizidine alkaloids in the relationship between larvae of Uresiphita reversalis (Lepidoptera: Pyralidae) and a host plant, Genista monspessulana. J. Chem. Ecol. 1990, 16, 1853–1865. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 13 June 2022).
- Bravo, L.E.; Muñoz, N. Epidemiology of cancer in Colombia. Colomb. Med. 2018, 49, 9–12. [Google Scholar] [CrossRef]
- Hernández, G. Cancer Epidemiology in Colombia: A Transition We Must Know. Medicina 2021, 43, 64–73. [Google Scholar]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New drugs, new toxicities: Severe side effects of modern targeted and immunotherapy of cancer and their management. Crit. Care 2017, 21, 89. [Google Scholar] [CrossRef] [Green Version]
- Nurgali, K.; Jagoe, R.T.; Abalo, R. Editorial: Adverse Effects of Cancer Chemotherapy: Anything New to Improve Tolerance and Reduce Sequelae? Front. Pharmacol. 2018, 9, 245. [Google Scholar] [CrossRef]
- Rigano, D.; Cardile, V.; Formisano, C.; Maldini, M.T.; Piacente, S.; Bevilacqua, J.; Russo, A.; Senatore, F. Genista sessilifolia DC. and Genista tinctoria L. inhibit UV light and nitric oxide-induced DNA damage and human melanoma cell growth. Chem. Biol. Interact. 2009, 180, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Bencherchar, I.; Demirtas, I.; Altun, M.; Gül, F.; Sarri, D.; Benayache, F.; Benayache, S.; Mekkiou, R. HPLC analysis, anti-oxidant activity of Genista ferox and its antiproliferative effect in HeLa cell line. Bangladesh J. Pharmacol. 2017, 12, 260–267. [Google Scholar] [CrossRef] [Green Version]
- Boutaghane, N.; Voutquenne-Nazabadioko, L.; Harakat, D.; Simon, A.; Kabouche, Z. Triterpene saponins of Genista ulicina Spach. Phytochemistry 2013, 93, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Lautié, E.; Russo, O.; Ducrot, P.; Boutin, J.A. Unraveling Plant Natural Chemical Diversity for Drug Discovery Purposes. Front. Pharmacol. 2020, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Caballero, J.M.; Cuca-Suárez, L.E.; Coy-Barrera, E. Bio-guided fractionation of ethanol extract of leaves of Esenbeckia alata Kunt (Rutaceae) led to the isolation of two cytotoxic quinoline alkaloids: Evidence of selectivity against leukemia cells. Biomolecules 2019, 9, 585. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Luo, J.; Qin, J.; Yang, M. Screening techniques for the identification of bioactive compounds in natural products. J. Pharm. Biomed. Anal. 2019, 168, 189–200. [Google Scholar] [CrossRef]
- Ory, L.; Nazih, E.-H.; Daoud, S.; Mocquard, J.; Bourjot, M.; Margueritte, L.; Delsuc, M.-A.; Bard, J.-M.; Pouchus, Y.F.; Bertrand, S.; et al. Targeting bioactive compounds in natural extracts—Development of a comprehensive workflow combining chemical and biological data. Anal. Chim. Acta 2019, 1070, 29–42. [Google Scholar] [CrossRef]
- Cárdenas-Laverde, D.; Barbosa-Cornelio, R.; Coy-Barrera, E. Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives. Plants 2021, 10, 2563. [Google Scholar] [CrossRef]
- Kellogg, J.J.; Wallace, E.D.; Graf, T.N.; Oberlies, N.H.; Cech, N.B. Conventional and accelerated-solvent extractions of green tea (Camellia sinensis) for metabolomics-based chemometrics. J. Pharm. Biomed. Anal. 2017, 145, 604–610. [Google Scholar] [CrossRef] [Green Version]
- D’Amelia, V.; Docimo, T.; Crocoll, C.; Rigano, M.M. Specialized Metabolites and Valuable Molecules in Crop and Medicinal Plants: The Evolution of Their Use and Strategies for Their Production. Genes 2021, 12, 936. [Google Scholar] [CrossRef]
- Weng, J.-K.; Lynch, J.H.; Matos, J.O.; Dudareva, N. Adaptive mechanisms of plant specialized metabolism connecting chemistry to function. Nat. Chem. Biol. 2021, 17, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, J.J.; Todd, D.A.; Egan, J.M.; Raja, H.A.; Oberlies, N.H.; Kvalheim, O.M.; Cech, N.B. Biochemometrics for natural products research: Comparison of data analysis approaches and application to identification of bioactive compounds. J. Nat. Prod. 2016, 79, 376–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cárdenas-Laverde, D.; Rincón-Aceldas, S.; Coy-Barrera, E. Identification of Antifungal Compounds from Piper Plants Against Fusarium oxysporum: An Untargeted Metabolite Profiling-Based Approach. Nat. Prod. Commun. 2022, 17, 1934578X221089995. [Google Scholar] [CrossRef]
- Caesar, L.K.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Opportunities and limitations for untargeted mass spectrometry metabolomics to identify biologically active constituents in complex natural product mixtures. J. Nat. Prod. 2019, 82, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Solorza-Bejarano, J.H. Regeneration pattern of the seedling of Genista monspessulana (L.) L.A.S. Johnson, in two ecological restoration scenarios. Colomb. For. 2017, 20, 131–143. [Google Scholar] [CrossRef]
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Ramawat, K.G.; Goyal, S. Co-evolution of Secondary Metabolites During Biological Competition for Survival and Advantage: An Overview BT—Co-Evolution of Secondary Metabolites. In Co-Evolution of Secondary Metabolites; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 3–17. ISBN 978-3-319-96397-6. [Google Scholar]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [Green Version]
- de Albergaria, E.T.; de Oliveira, A.F.M.; Albuquerque, U.P. Effect of rainfall and soil fertility on total phenol and tannin contents in Cenostigma microphyllum (Mart. ex G. Don) E. Gagnon & G.P. Lewis (Fabaceae). Acta Physiol. Plant. 2021, 43, 61. [Google Scholar]
- Albrecht, C.F.; Stander, M.A.; Grobbelaar, M.C.; Colling, J.; Kossmann, J.; Hills, P.N.; Makunga, N.P. LC–MS-based metabolomics assists with quality assessment and traceability of wild and cultivated plants of Sutherlandia frutescens (Fabaceae). S. Afr. J. Bot. 2012, 82, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Zhu, C.-S.; Lin, Z.-J.; Xiao, M.-L.; Niu, H.-J.; Zhang, B. The spectrum-effect relationship—a rational approach to screening effective compounds, reflecting the internal quality of Chinese herbal medicine. Chin. J. Nat. Med. 2016, 14, 177–184. [Google Scholar] [CrossRef]
- MacCallum, R.C.; Zhang, S.; Preacher, K.J.; Rucker, D.D. On the practice of dichotomization of quantitative variables. Psychol. Methods 2002, 7, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Royston, P. The cost of dichotomising continuous variables. BMJ 2006, 332, 1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, E.R.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Biochemometrics to identify synergists and additives from botanical medicines: A case study with Hydrastis canadensis (goldenseal). J. Nat. Prod. 2018, 81, 484–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, T.; Takahashi, R.; Namiki, H. Syntheses of (+)-Cytisine, (−)-Kuraramine, (−)-Isokuraramine, and (−)-Jussiaeiine A. J. Org. Chem. 2005, 70, 499–504. [Google Scholar] [CrossRef]
- Huang, K.-F.; Yen, Y.-F. Three Prenylated Isoflavones from Erythrina variegate. J. Chin. Chem. Soc. 1996, 43, 515–518. [Google Scholar] [CrossRef]
- Kolanoś, R.; Wysocka, W.; Brukwicki, T. A comparative study of NMR chemical shifts of sparteine thiolactams and lactams. Tetrahedron 2003, 59, 5531–5537. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, L.; Jia, Z.; Chen, J. Stereochemical study of anagyrine-type quinolizidine alkaloids by 15N and 2D NMR spectroscopy. Magn. Reson. Chem. 1992, 30, 511–514. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Li, J.-S.; Jiang, Z.-R.; Kubo, H.; Higashiyama, K.; Ohmiya, S. Lupin Alkaloids from Chinese Maackia amurensis. Chem. Pharm. Bull. 2000, 48, 641–645. [Google Scholar] [CrossRef] [Green Version]
- Lane, G.A.; Newman, R.H. Isoflavones from Lupinus angustifolius root. Phytochemistry 1986, 26, 295–300. [Google Scholar] [CrossRef]
- Al-Azizi, M.M.; Al-Said, M.S.; El-Olemy, M.M.; Sattar, E.A.; Khalifa, A.S. Rhombifoline and 5,6-dehydrolupanine from Anagyrus foetida L. Arch. Pharm. Res. 1994, 17, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Gotti, C.; Clementi, F. Cytisine and cytisine derivatives. More than smoking cessation aids. Pharmacol. Res. 2021, 170, 105700. [Google Scholar] [CrossRef]
- Yu, L.; Wang, X.; Chen, Z.-F.; Jiang, B.; Shang, D.-Y.; Sun, Y.-X.; Yang, J.-H.; Zhang, L.-F.; Ji, Y.-B. Cytisine induces apoptosis of HepG2 cells. Mol. Med. Rep. 2017, 16, 3363–3370. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.-T.; Li, T.-Z.; Li, S.-M.; Wang, C.; Wang, H.; Luo, Y.-H.; Piao, X.-J.; Wang, J.-R.; Zhang, Y.; Zhang, T.; et al. Cytisine exerts anti-tumour effects on lung cancer cells by modulating reactive oxygen species-mediated signalling pathways. Artif. Cells Nanomed. Biotechnol. 2020, 48, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Petruczynik, A.; Wróblewski, K.; Misiurek, J.; Plech, T.; Szalast, K.; Wojtanowski, K.; Mroczek, T.; Szymczak, G.; Waksmundzka-Hajnos, M.; Tutka, P. Determination of Cytisine and N-Methylcytisine from Selected Plant Extracts by High-Performance Liquid Chromatography and Comparison of Their Cytotoxic Activity. Toxins 2020, 12, 557. [Google Scholar] [CrossRef] [PubMed]
- Ateba, S.B.; Mvondo, M.A.; Djiogue, S.; Zingué, S.; Krenn, L.; Njamen, D. A Pharmacological Overview of Alpinumisoflavone, a Natural Prenylated Isoflavonoid. Front. Pharmacol. 2019, 10, 952. [Google Scholar] [CrossRef]
- Nana, F.; Sandjo, L.P.; Keumedjio, F.; Ambassa, P.; Malik, R.; Kuete, V.; Rincheval, V.; Choudhary, M.I.; Ngadjui, B.T. Ceramides and cytotoxic constituents from Ficus glumosa Del. (Moraceae). J. Braz. Chem. Soc. 2012, 23, 482–487. [Google Scholar] [CrossRef] [Green Version]
- Innocenti, G.; Dall’Acqua, S.; Viola, G.; Loi, M.C. Cytotoxic constituents from Anagyris foetida leaves. Fitoterapia 2006, 77, 595–597. [Google Scholar] [CrossRef]
- Gao, L.; Wang, K.; Zhou, Y.; Fang, J.; Qin, X.; Du, G. Uncovering the anticancer mechanism of Compound Kushen Injection against HCC by integrating quantitative analysis, network analysis and experimental validation. Sci. Rep. 2018, 8, 624. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Pathania, A.S.; Saxena, A.K.; Vishwakarma, R.A.; Ali, A.; Bhushan, S. The anticancer potential of flavonoids isolated from the stem bark of Erythrina suberosa through induction of apoptosis and inhibition of STAT signaling pathway in human leukemia HL-60 cells. Chem. Biol. Interact. 2013, 205, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frasinyuk, M.S.; Zhang, W.; Wyrebek, P.; Yu, T.; Xu, X.; Sviripa, V.M.; Bondarenko, S.P.; Xie, Y.; Ngo, H.X.; Morris, A.J.; et al. Developing antineoplastic agents that target peroxisomal enzymes: Cytisine-linked isoflavonoids as inhibitors of hydroxysteroid 17-beta-dehydrogenase-4 (HSD17B4). Org. Biomol. Chem. 2017, 15, 7623–7629. [Google Scholar] [CrossRef] [PubMed]
- Cardellicchio, C.; Capozzi, M.A.M.; Naso, F. The Betti base: The awakening of a sleeping beauty. Tetrahedron Asymmetry 2010, 21, 507–517. [Google Scholar] [CrossRef]
- Peng, T.-T.; Sun, X.-R.; Liu, R.-H.; Hua, L.-X.; Cheng, D.-P.; Mao, B.; Li, X.-N. Cytisine-Pterocarpan Derived Compounds: Biomimetic Synthesis and Apoptosis-Inducing Activity in Human Breast Cancer Cells. Molecules 2018, 23, 3059. [Google Scholar] [CrossRef] [Green Version]
- Balachandran, C.; Duraipandiyan, V.; Arun, Y.; Sangeetha, B.; Emi, N.; Al-Dhabi, N.A.; Ignacimuthu, S.; Inaguma, Y.; Okamoto, A.; Perumal, P.T. Isolation and characterization of 2-hydroxy-9,10-anthraquinone from Streptomyces olivochromogenes (ERINLG-261) with antimicrobial and antiproliferative properties. Rev. Bras. Farmacogn. 2016, 26, 285–295. [Google Scholar] [CrossRef] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]




| PC-3 b | SiHa b | A549 b | L929 c | |||||
|---|---|---|---|---|---|---|---|---|
| Samples a | IC50 c | CI d | IC50 c | CI d | IC50 c | CI d | IC50 c | CI d |
| Gm1 | 151 | 141–165 | 136 | 124–151 | >500 | - | >500 | - |
| Gm2 | 119 | 107–131 | 271 | 246–290 | >500 | - | 313 | 291–328 |
| Gm3 | 57.8 | 52.6–63.0 | 75.1 | 69.8–83.4 | >500 | - | >500 | - |
| Gm4 | 196 | 186–210 | 216 | 194–233 | >500 | - | 389 | 369–416 |
| Gm5 | 42.8 | 39.4–47.1 | 61.4 | 57.1–68.2 | >500 | - | >500 | - |
| Gm6 | 211 | 190–228 | 285 | 271–299 | >500 | - | 226 | 206–251 |
| Gm7 | 55.6 | 50.6–61.2 | 77.8 | 70.8–84.0 | >500 | - | >500 | - |
| Gm8 | 92.8 | 87.2–99.3 | 258 | 245–276 | >500 | - | 390 | 362–424 |
| Gm9 | 31.6 | 29.7–33.5 | 48.4 | 43.6–53.2 | >500 | - | >500 | - |
| Gm10 | 26.3 | 24.2–29.2 | 60.0 | 56.4–63.6 | >500 | - | >500 | - |
| PC-3 b | SiHa b | A549 b | L929 c | |||||
|---|---|---|---|---|---|---|---|---|
| Samples a | IC50 c | CI d | IC50 c | CI d | IC50 c | CI d | IC50 c | CI d |
| 1 | 15.8 | 14.7–16.6 | 357 | 341–388 | 42.5 | 39.5–45.5 | 102.7 | 97.6–107 |
| 2 | 18.6 | 17.9–20.1 | 19.6 | 17.6–20.6 | >303 | - | >297 | - |
| 3 | 21.3 | 19.2–23.2 | >403 | - | >403 | - | 22.3 | 19.8–25 |
| 4 | 33.5 | 31.8–35.8 | >409 | - | >409 | - | 36.5 | 33.6–38.7 |
| 5 | 28.4 | 26.4–31.5 | >490 | - | 36.3 | 34.5–40.7 | 91.3 | 85.8–95 |
| 6 | 34.3 | 32.2–35.7 | >296 | - | >296 | - | >296 | - |
| 7 | 31.5 | 29.6–33.1 | >406 | - | >406 | - | 85.9 | 79–95.3 |
| curcumin | 9.5 | 8.8–10.6 | 6.5 | 6.2–6.9 | 8.4 | 7.7–8.7 | 104.8 | 93.3–116 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz, L.; Cely-Veloza, W.; Coy-Barrera, E. Identification of Anti-Proliferative Compounds from Genista monspessulana Seeds through Covariate-Based Integration of Chemical Fingerprints and Bioactivity Datasets. Molecules 2022, 27, 3996. https://doi.org/10.3390/molecules27133996
Díaz L, Cely-Veloza W, Coy-Barrera E. Identification of Anti-Proliferative Compounds from Genista monspessulana Seeds through Covariate-Based Integration of Chemical Fingerprints and Bioactivity Datasets. Molecules. 2022; 27(13):3996. https://doi.org/10.3390/molecules27133996
Chicago/Turabian StyleDíaz, Luis, Willy Cely-Veloza, and Ericsson Coy-Barrera. 2022. "Identification of Anti-Proliferative Compounds from Genista monspessulana Seeds through Covariate-Based Integration of Chemical Fingerprints and Bioactivity Datasets" Molecules 27, no. 13: 3996. https://doi.org/10.3390/molecules27133996