Computational Study of Asian Propolis Compounds as Potential Anti-Type 2 Diabetes Mellitus Agents by Using Inverse Virtual Screening with the DIA-DB Web Server, Tanimoto Similarity Analysis, and Molecular Dynamic Simulation
Abstract
:1. Introduction
2. Results
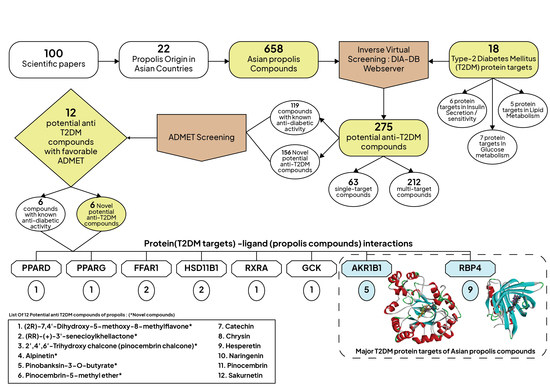
2.1. Inverse Virtual Screening and Identification of Propolis Compounds with Potential Anti-T2DM Activity
| Mode of Action | Protein Target | Function | PDB Code | Percentage of Potential Compounds (Total of Potential Compounds/Total of Test Compounds) | Test Compounds with the Lowest Energy (Kcal/mol) | Test Compound Name with the Lowest Binding Energy |
|---|---|---|---|---|---|---|
| Regulation of insulin secretion and sensitivity | DPP4 | Cleaves and inactivates glucagon-like peptide-1 that stimulates insulin secretion and inhibits glucagon secretion [118,119] | 4A5S | 10.3% (68/658) | −10.90 | Retusapurpurin A |
| FFAR1 | G-protein-coupled receptors bind with a free fatty acid or its specific agonist to support beta-cell function for insulin secretion [120,121] | 4PHU | 11.1% (73/658) | −10.40 | TB1 and isonymphaeol-B | |
| HSD11B1 | Catalyzes the interconversion of inactive glucocorticoid (cortisone) to active glucocorticoid (cortisol) that stimulates insulin resistance [122,123] | 4K1L | 20.8% (137/658) | −11.90 | 23-hydroxymangiferonic acid | |
| INSR | Expressed in insulin-responsive cells and acts as the initial point of insulin signaling [124] | 3EKN | 0.9% (6/658) | −10.10 | Taraxasterol | |
| PTPN9 | A negative regulator of insulin signaling via catalyzing the rapid dephosphorylation of insulin receptor resulting in insulin resistance [124,125] | 4GE6 | 0.3% (2/658) | −9.40 | Garcinone B | |
| RBP4 | An adipocyte-secreted molecule that activates innate immune responses and induces inflammation resulting in insulin resistance [126] | 2WR6 | 12.5% (82/658) | −12.40 | 10-hydroxybenzo[j]fluoranthene | |
| Regulation of glucose metabolism | AKR1B1 | Catalyzes the reduction of glucose to sorbitol in the polyol pathway, thus contributing to diabetes complications [127,128] | 3G5E | 24.9% (164/658) | −11.30 | TB2 |
| AMY2A | Catalyzes the hydrolysis of the α-1,4-d-glycobsidic bond of starch to produce glucose [129] | 4GQR | 8.7% (57/658) | −10.90 | 24-(Z)-3-oxolanosta-1,7,24-trien-26-oic acid | |
| FBP1 | A major regulator that catalyzes glucose production in the second last step of gluconeogenesis [130] | 2JJK | 0% | - | ||
| GCK | Regulates glucose homeostasis and synthesizes glucose 6-phosphate from glucose in the glycolytic pathway [131] | 3IMX | 15% (98/658) | −10.40 | 6-cinnamylchrysin | |
| MGAM | Catalyzes the last step of starch digestion via hydrolysis of 1,4-α bonds in starch to produce glucose [132] | 3L4Y | 1.2% (8/658) | −9.80 | 24-(Z)-3-oxolanosta-1,7,24-trien-26-oic acid | |
| PDK2 | Inhibits pyruvate dehydrogenase activity through a phosphorylation reaction which causes glycolytic disruption and glucose oxidation [131] | 4MPC | 0.8% (5/658) | −9.30 | 24-(Z)-3-oxolanosta-1,7,24-trien-26-oic acid | |
| PYGL | Catalyze the phosphorolysis of α-1,4-glycosidic bonds in glycogen to glucose-1-phosphat [133] | 3DDS | 0.6% (4/658) | −9.60 | Acetoxymangiferonic acid | |
| Regulation of lipid metabolism | NR5A2 | Regulates the expression of genes involved in steroidogenesis, bile acid metabolism, and cholesterol synthesis [134] | 4DOR | 0.5% (3/658) | −9.70 | Retusapurpurin A |
| PPARA | Regulates the expression of genes involved in uptake, binding, and oxidation of fatty acids in the liver as well as lipoprotein assembly and lipid transport [135] | 3FEI | 1.2% (8/658) | −9.50 | Isonymphaeol-B | |
| PPARD | Regulates fatty acid catabolism in skeletal muscle [135] | 3PEQ | 18.4% (121/658) | −11.80 | Amphidinolide X | |
| PPARG | Regulates the expression of genes involved in adipogenesis and lipid metabolism, particularly fatty acid transport, lipid droplet formation, triacylglycerol metabolism, as well as lipolysis of triglycerides [135] | 2FVJ | 13.2% (87/658) | −12.00 | Retusapurpurin A | |
| RXRA | Mediates gene transcription by forming heterodimers with PPAR [136] | 1FM9 | 9.9% (65/658) | −12.00 | Lespeol |
2.2. Molecular Similarity Evaluation of Selected Propolis Compounds with Approved Anti-T2DM Drugs
2.3. Summary of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Parameters of Selected Propolis Compounds
2.4. Analyzing the Molecular Docking Simulation Results of Asian Propolis Compounds with RBP4 and AKR1B1
2.5. Molecular Dynamic Simulations of Selected Asian Propolis Compounds
3. Materials and Methods
3.1. Collection of All Propolis Compounds in Asian Countries from All of the Previous Reports Related to Asian Propolis
3.2. Preparation of Compounds Structure and Inverse Virtual Screening of Potential Anti-T2DM Agents and Novel Compounds Analysis
3.3. Similarity Studies of Potential Compounds with Approved Anti-T2DM Drugs
3.4. Studies on Oral Bioavailability and Absorption, Distribution, Excretion, and Toxicity (ADMET) Properties of All Potential Compounds
3.5. Molecular Interaction Analysis among T2DM Targets and Potential Compounds of Propolis with Favorable ADMET Properties
3.6. Molecular Dynamic Simulation of Selected Compounds of Asian Propolis to Two Major Diabetes Targets
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ralph, A.D.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.; Kahn, C.; Raz, I.; Shulman, G. Type 2 diabetes mellitus. Nat. Rev. Dis. Prim. 2015, 1, 15019. [Google Scholar]
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.; Müller, M.J. Type 2 diabetes in Asia: Where do we go from here? Eur. J. Clin. Nutr. 2017, 71, 801–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhury, A.; Duvoor, C.; Reddy Dendi, V.S.; Kraleti, S.; Chada, A.; Ravilla, R.; Marco, A.; Shekhawat, N.S.; Montales, M.T.; Kuriakose, K.; et al. Clinical Review of Antidiabetic Drugs: Implications for Type 2 Diabetes Mellitus Management. Front. Endocrinol. 2017, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Pang, G.-M.; Li, F.-X.; Yan, Y.; Zhang, Y.; Kong, L.-L.; Zhu, P.; Wang, K.-F.; Zhang, F.; Liu, B.; Lu, C. Herbal medicine in the treatment of patients with type 2 diabetes mellitus. Chin. Med. J. (Engl.) 2019, 132, 78–85. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Dai, Y.; Peng, J. Natural products for the treatment of type 2 diabetes mellitus: Pharmacology and mechanisms. Pharmacol. Res. 2018, 130, 451–465. [Google Scholar] [CrossRef]
- Yusuf, A.P.; Zhang, J.Y.; Li, J.Q.; Muhammad, A.; Abubakar, M.B. Herbal medications and natural products for patients with covid-19 and diabetes mellitus: Potentials and challenges. Phytomed. Plus 2022, 2, 100280. [Google Scholar] [CrossRef]
- Braakhuis, A. Evidence on the health benefits of supplemental propolis. Nutrients 2019, 11, 2075. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Zhang, C.-P.; Wang, K.; Li, G.Q.; Hu, F.-L. Recent advances in the chemical composition of propolis. Molecules 2014, 19, 19610–19632. [Google Scholar] [CrossRef] [Green Version]
- Siheri, W.; Alenezi, S.; Tusiimire, J.; Watson, D.G. Bee Products-Chemical and Biological Properties. JB Metzler 2017, 7, 137–178. [Google Scholar]
- Samadi, N.; Mozaffari-Khosravi, H.; Rahmanian, M.; Askarishahi, M. Effects of bee propolis supplementation on glycemic control, lipid profile and insulin resistance indices in patients with type 2 diabetes: A randomized, double-blind clinical trial. J. Integr. Med. 2017, 15, 124–134. [Google Scholar] [CrossRef]
- Zakerkish, M.; Jenabi, M.; Zaeemzadeh, N.; Hemmati, A.A.; Neisi, N. The Effect of Iranian Propolis on Glucose Metabolism, Lipid Profile, Insulin Resistance, Renal Function and Inflammatory Biomarkers in Patients with Type 2 Diabetes Mellitus: A Randomized Double-Blind Clinical Trial. Sci. Rep. 2019, 9, 7289. [Google Scholar] [CrossRef] [PubMed]
- El Adaouia Taleb, R.; Djebli, N.; Chenini, H.; Sahin, H.; Kolayli, S. In vivo and in vitro anti-diabetic activity of ethanolic propolis extract. J. Food Biochem. 2020, 44, e13267. [Google Scholar] [CrossRef] [PubMed]
- El Rabey, H.A.; Al-Seeni, M.N.; Bakhashwain, A.S. The antidiabetic activity of Nigella sativa and propolis on streptozotocin-induced diabetes and diabetic nephropathy in male rats. Evid.-Based Complement. Altern. Med. 2017, 2017, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Li, Y.; Chen, M.; Xuan, H. Effects of encapsulated propolis on blood glycemic control, lipid metabolism, and insulin resistance in type 2 diabetes mellitus rats. Evid.-Based Complement. Altern. Med. 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Karimian, J.; Hadi, A.; Pourmasoumi, M.; Najafgholizadeh, A.; Ghavami, A. The efficacy of propolis on markers of glycemic control in adults with type 2 diabetes mellitus: A systematic review and meta-analysis. Phyther. Res. 2019, 33, 1616–1626. [Google Scholar] [CrossRef]
- El-Sharkawy, H.M.; Anees, M.M.; Van Dyke, T.E. Propolis Improves Periodontal Status and Glycemic Control in Patients With Type 2 Diabetes Mellitus and Chronic Periodontitis: A Randomized Clinical Trial. J. Periodontol. 2016, 87, 1418–1426. [Google Scholar] [CrossRef]
- Sánchez-Pérez, A.; Muñoz, A.; Peña-García, J.; den-Haan, H.; Bekas, N.; Katsikoudi, A.; Tzakos, A.G.; Péréz-Sánchez, H. DIA-DB: A web-accessible database for the prediction of diabetes drugs. Lect. Notes Comput. Sci. (Incl. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2015, 9044, 655–663. [Google Scholar] [CrossRef]
- Perez-Sanchez, H.; Den-Haan, H.; Peña-García, J.; Lozano-Sánchez, J.; Martinez Moreno, M.E.; Sánchez-Pérez, A.; Munoz, A.; Ruiz-Espinosa, P.; Pereira, A.S.P.; Katsikoudi, A. DIA-DB: A database and web server for the prediction of diabetes drugs. J. Chem. Inf. Model. 2020, 60, 4124–4130. [Google Scholar] [CrossRef]
- Pereira, A.S.P.; Haan, H.D.; Peña-García, J.; Moreno, M.M.; Pérez-Sánchez, H.; Apostolides, Z. Exploring african medicinal plants for potential anti-diabetic compounds with the DIA-DB inverse virtual screening web server. Molecules 2019, 24, 2002. [Google Scholar] [CrossRef] [Green Version]
- Tanvir, E.M.; Hasan, M.A.; Nayan, S.I.; Islam, T.; Ahmed, T.; Hossen, M.S.; Perveen, R.; Rahman, S.; Afroz, R.; Afroz, R.; et al. Ameliorative effects of ethanolic constituents of Bangladeshi propolis against tetracycline-induced hepatic and renal toxicity in rats. J. Food Biochem. 2019, 43, e12958. [Google Scholar] [CrossRef] [PubMed]
- Ristivojević, P.; Stević, T.; Starović, M.; Pavlović, S.; Özcan, M.M.; Berić, T.; Dimkić, I. Phenolic composition and biological activities of geographically different type of propolis and black cottonwood resins against oral streptococci, vaginal microbiota and phytopathogenic Fusarium species. J. Appl. Microbiol. 2020, 129, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Chen, Y.; Zhang, J.; You, M.; Wang, K.; Hu, F. Phytomedicine Mechanisms underlying the wound healing potential of propolis based on its in vitro antioxidant activity. Phytomedicine 2017, 34, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Brunori, F.; Tulimieri, A.; Piccinino, D.; Meschini, R.; Saladino, R. Laccase-Mediated Enhancement of the Antioxidant Activity of Propolis and Poplar Bud Exudates. ACS Omega 2017, 2, 2515–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.; Xuan, H.; Sun, A.; Liu, R.; Cui, J. Preparative separation of polyphenols from water-soluble fraction of Chinese propolis using macroporous absorptive resin coupled with preparative high performance liquid chromatography. J. Chromatogr. B 2016, 1012, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shen, X.; Wang, K.; Cao, X.; Zhang, C.; Zheng, H.; Hu, F. Antioxidant activities and molecular mechanisms of the ethanol extracts of Baccharis propolis and Eucalyptus propolis in RAW64.7 cells. Pharm. Biol. 2016, 54, 2220–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, H.; Wang, Y.; Li, A.; Fu, C.; Wang, Y.; Peng, W. Bioactive Components of Chinese Propolis Water Extract on Antitumor Activity and Quality Control. Evid. Based Complement. Altern. Med. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Sun, H.; Zhang, J.; Zhang, X.; Zhao, L.; Guo, S.; Li, Y.; Jiao, P.; Wang, H.; Qin, S.; et al. Ethanol extract of propolis protects macrophages from oxidized low density lipoprotein-induced apoptosis by inhibiting CD36 expression and endoplasmic reticulum stress-C/EBP homologous protein pathway. BMC Complement. Altern. Med. 2015, 2015, 230. [Google Scholar] [CrossRef] [Green Version]
- Cui-ping, Z.; Shuai, H.; Wen-ting, W.; Shun, P.; Xiao-ge, S.; Ya-jing, L.; Fu-liang, H. Development of high-performance liquid chromatographic for quality and authenticity control of Chinese propolis. J. Food Sci. 2014, 79, C1315–C1322. [Google Scholar] [CrossRef]
- Wang, K.; Ping, S.; Huang, S.; Hu, L.; Xuan, H.; Zhang, C.; Hu, F. Molecular mechanisms underlying the in vitro anti-inflammatory effects of a flavonoid-rich ethanol extract from chinese propolis (poplar type). Evid. Based Complement. Altern. Med. 2013, 2013, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Nie, P.; Xia, Z.; Sun, D.W.; He, Y. Application of visible and near infrared spectroscopy for rapid analysis of chrysin and galangin in chinese propolis. Sensors 2013, 13, 10539–10549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Yang, H.; Zhang, X.; Yu, L. Identification and quantification of phytochemical composition and anti-inflammatory and radical scavenging properties of methanolic extracts of Chinese propolis. J. Agric. Food Chem. 2012, 60, 12403–12410. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, Q.; Wang, M.; Zhang, L. Metabolomics reveals discrimination of Chinese propolis from different climatic regions. Foods 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Dong, Y.; Du, H.; Shi, H.; Peng, Y.; Li, X. Antioxidant compounds from propolis collected in Anhui, China. Molecules 2011, 16, 3444–3455. [Google Scholar] [CrossRef]
- Yang, C.; Luo, L.; Zhang, H.; Yang, X.; Lv, Y.; Song, H. Common aroma-active components of propolis from 23 regions of China. J. Sci. Food Agric. 2010, 90, 1268–1282. [Google Scholar] [CrossRef]
- Sha, N.; Guan, S.-H.; Lu, Z.-Q.; Chen, G.-T.; Huang, H.-L.; Xie, F.-B.; Yue, Q.-X.; Liu, X.; Guo, D.-A. Cytotoxic constituents of Chinese propolis. J. Nat. Prod. 2009, 72, 799–801. [Google Scholar] [CrossRef]
- Sha, N.; Huang, H.-L.; Zhang, J.-Q.; Chen, G.-T.; Tao, S.-J.; Yang, M.; Li, X.-N.; Li, P.; Guo, D.-A. Simultaneous quantification of eight major bioactive phenolic compounds in Chinese propolis by high-performance liquid chromatography. Nat. Prod. Commun. 2009, 4, 1934578X0900400615. [Google Scholar] [CrossRef] [Green Version]
- Usia, T.; Banskota, A.H.; Tezuka, Y.; Midorikawa, K.; Matsushige, K.; Kadota, S. Constituents of Chinese propolis and their antiproliferative activities. J. Nat. Prod. 2002, 65, 673–676. [Google Scholar] [CrossRef]
- El-Guendouz, S.; Lyoussi, B.; Miguel, M.G. Insight on Propolis from Mediterranean Countries: Chemical Composition, Biological Activities and Application Fields. Chem. Biodivers. 2019, 16, e1900094. [Google Scholar] [CrossRef]
- Kasote, D.M.; Pawar, M.V.; Bhatia, R.S.; Nandre, V.S.; Gundu, S.S.; Jagtap, S.D.; Kulkarni, M.V. HPLC, NMR based chemical profiling and biological characterisation of Indian propolis. Fitoterapia 2017, 122, 52–60. [Google Scholar] [CrossRef]
- Sadhana, N.; Lohidasan, S.; Mahadik, K.R. Marker-based standardization and investigation of nutraceutical potential of Indian propolis. J. Integr. Med. 2017, 15, 483–494. [Google Scholar] [CrossRef]
- Choudhari, M.K.; Punekar, S.A.; Ranade, R.V.; Paknikar, K.M. Antimicrobial activity of stingless bee (Trigona sp.) propolis used in the folk medicine of Western Maharashtra, India. J. Ethnopharmacol. 2012, 141, 363–367. [Google Scholar] [CrossRef]
- Naik, D.G.; Vaidya, H.S.; Namjoshi, T.P. Essential Oil of Indian Propolis: Chemical Composition and Repellency against the Honeybee Apis florea. Chem. Biodivers. 2013, 10, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yuan, X.; Pineda, M.; Liang, Z.; He, J.; Sun, S.; Pan, T.; Li, K. A comparative study between Chinese propolis and Brazilian green propolis: Metabolite profile and bioactivity. Food Funct. 2020, 11, 2368–2379. [Google Scholar] [CrossRef] [PubMed]
- Miyata, R.; Sahlan, M.; Ishikawa, Y.; Hashimoto, H.; Honda, S.; Kumazawa, S. Propolis Components from Stingless Bees Collected on South Sulawesi, Indonesia, and Their Xanthine Oxidase Inhibitory Activity. J. Nat. Prod. 2019, 82, 205–2010. [Google Scholar] [CrossRef]
- Trusheva, B.; Todorov, I.; Ninova, M.; Najdenski, H.; Daneshmand, A.; Bankova, V. Antibacterial mono-and sesquiterpene esters of benzoic acids from Iranian propolis. Chem. Cent. J. 2010, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Dinparast-Djadid, N.; Frankland, A.C.H.; Saleh Azizian, M.D. Chemoprotection of MNNG-initiated gastric cancer in rats using Iranian propolis. Arch. Iran. Med. 2015, 18, 18. [Google Scholar]
- Asgharpur, F.; Moghadamnia, A.A.; Kazemi, S.; Nouri, H.R.; Motallebnejad, M. Applying GC-MS analysis to identify chemical composition of Iranian propolis prepared with different solvent and evaluation of its biological activity. Casp. J. Intern. Med. 2020, 11, 191–198. [Google Scholar] [CrossRef]
- Bazmandegan, G.; Boroushaki, M.T.; Shamsizadeh, A.; Ayoobi, F.; Hakimizadeh, E.; Allahtavakoli, M. Brown propolis attenuates cerebral ischemia-induced oxidative damage via affecting antioxidant enzyme system in mice. Biomed. Pharmacother. 2017, 85, 503–510. [Google Scholar] [CrossRef]
- Mohammadzadeh, S.; Shariatpanahi, M.; Hamedi, M.; Ahmadkhaniha, R.; Samadi, N.; Ostad, S.N. Chemical composition, oral toxicity and antimicrobial activity of Iranian propolis. Food Chem. 2007, 103, 1097–1103. [Google Scholar] [CrossRef]
- Tukmechi, A.; Ownagh, A.; Mohebbat, A. In vitro antibacterial activities of ethanol extract of Iranian propolis (EEIP) against fish pathogenic bacteria (Aeromonas hydrophila, Yersinia ruckeri & Streptococcus iniae). Braz. J. Microbiol. 2010, 41, 1086–1092. [Google Scholar] [PubMed]
- Sulaiman, G.M.; Sammarrae, K.W.A.; Ad’hiah, A.H.; Zucchetti, M.; Frapolli, R.; Bello, E.; Erba, E.; D’Incalci, M.; Bagnati, R. Chemical characterization of iraqi propolis samples and assessing their antioxidant potentials. Food Chem. Toxicol. 2011, 49, 2415–2421. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, S.; Ueda, R.; Hamasaka, T.; Fukumoto, S.; Fujimoto, T.; Nakayama, T. Antioxidant prenylated flavonoids from propolis collected in Okinawa, Japan. J. Agric. Food Chem. 2007, 55, 7722–7725. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, S.; Nakamura, J.; Murase, M.; Miyagawa, M.; Ahn, M.R.; Fukumoto, S. Plant origin of Okinawan propolis: Honeybee behavior observation and phytochemical analysis. Naturwissenschaften 2008, 95, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Luo, L.; Cui, M.; Hao, Y.; Liu, T.; Huang, X.; Guo, X. Chemical Composition and Antioxidant Activity of Essential Oil of Chinese Propolis. Chem. Biodivers. 2020, 17, e1900489. [Google Scholar] [CrossRef]
- Shaheen, S.A.; Zarga, M.H.A.; Nazer, I.K.; Darwish, R.M.; Al-Jaber, H.I. Chemical constituents of Jordanian propolis. Nat. Prod. Res. 2011, 25, 1312–1318. [Google Scholar] [CrossRef]
- Abutaha, N. Apoptotic Potential and Chemical Composition of Jordanian Propolis Extract against Different Cancer Cell Lines. J. Microbiol. Biotechnol. 2019, 30, 893–902. [Google Scholar] [CrossRef]
- Noureddine, H.; Hage-Sleiman, R.; Wehbi, B.; Fayyad-Kazan, A.H.; Hayar, S.; Traboulssi, M.; Alyamani, O.A.; Faour, W.H.; ElMakhour, Y. Chemical characterization and cytotoxic activity evaluation of Lebanese propolis. Biomed. Pharmacother. 2017, 95, 298–307. [Google Scholar] [CrossRef]
- Ong, T.H.; Chitra, E.; Ramamurthy, S.; Siddalingam, R.P.; Yuen, K.H.; Ambu, S.P.; Davamani, F. Chitosan-propolis nanoparticle formulation demonstrates anti-bacterial activity against Enterococcus faecalis biofilms. PLoS ONE 2017, 12, e0174888. [Google Scholar] [CrossRef] [Green Version]
- Usman, U.Z.; Bakar, A.B.A.; Mohamed, M. Phytochemical composition and activity against hyperglycaemia of Malaysian propolis in diabetic rats. Biomed. Res. 2016, 27, 46–51. [Google Scholar]
- Li, F.; Awale, S.; Zhang, H.; Tezuka, Y.; Esumi, H.; Kadota, S. Chemical constituents of propolis from Myanmar and their preferential cytotoxicity against a human pancreatic cancer cell line. J. Nat. Prod. 2009, 72, 1283–1287. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Awale, S.; Tezuka, Y.; Kadota, S. Cytotoxic constituents of propolis from Myanmar and their structure–activity relationship. Biol. Pharm. Bull. 2009, 32, 2075–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awale, S.; Shrestha, S.P.; Tezuka, Y.; Ueda, J.Y.; Matsushige, K.; Kadota, S. Neoflavonoids and related constituents from nepalese propolis and their nitric oxide production inhibitory activity. J. Nat. Prod. 2005, 68, 858–864. [Google Scholar] [CrossRef]
- Funakoshi-Tago, M.; Okamoto, K.; Izumi, R.; Tago, K.; Yanagisawa, K.; Narukawa, Y.; Kiuchi, F.; Kasahara, T.; Tamura, H. Anti-inflammatory activity of flavonoids in Nepalese propolis is attributed to inhibition of the IL-33 signaling pathway. Int. Immunopharmacol. 2015, 25, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi-Tago, M.; Ohsawa, K.; Ishikawa, T.; Nakamura, F.; Ueda, F.; Narukawa, Y.; Kiuchi, F.; Tamura, H.; Tago, K.; Kasahara, T. Inhibitory effects of flavonoids extracted from Nepalese propolis on the LPS signaling pathway. Int. Immunopharmacol. 2016, 40, 550–560. [Google Scholar] [CrossRef]
- Jiang, X.; Tian, J.; Zheng, Y.; Zhang, Y.; Wu, Y.; Zhang, C.; Zheng, H.; Hu, F. A new propolis type from changbai mountains in north-east china: Chemical composition, botanical origin and biological activity. Molecules 2019, 24, 1369. [Google Scholar] [CrossRef] [Green Version]
- Okińczyc, P.; Paluch, E.; Franiczek, R.; Widelski, J.; Wojtanowski, K.K.; Mroczek, T.; Krzyżanowska, B.; Skalicka-Woźniak, K.; Sroka, Z. Antimicrobial activity of Apis mellifera L. and Trigona sp. propolis from Nepal and its phytochemical analysis. Biomed. Pharmacother. 2020, 129, 110435. [Google Scholar] [CrossRef]
- Shrestha, S.P.; Narukawa, Y.; Takeda, T. Chemical constituents of Nepalese propolis (II). Chem. Pharm. Bull. 2007, 55, 926–929. [Google Scholar] [CrossRef] [Green Version]
- Popova, M.; Dimitrova, R.; Al-Lawati, H.T.; Tsvetkova, I.; Najdenski, H.; Bankova, V. Omani propolis: Chemical profiling, antibacterial activity and new propolis plant sources. Chem. Cent. J. 2013, 7, 158. [Google Scholar] [CrossRef] [Green Version]
- Desamero, M.J.; Kakuta, S.; Tang, Y.; Chambers, J.K.; Uchida, K.; Estacio, M.A.; Cervancia, C.; Kominami, Y.; Ushio, H.; Nakayama, J.; et al. Tumor-suppressing potential of stingless bee propolis in in vitro and in vivo models of differentiated-type gastric adenocarcinoma. Sci. Rep. 2019, 9, 19635. [Google Scholar] [CrossRef]
- Ragasa, C.Y.; Galian, R.A.F.; Ebajo, V.D.; Aguda, R.M.; Cervancia, C.R.; Shen, C.C. Propolins and glyasperin a from stingless bee nests. Rev. Bras. Farmacogn. 2015, 25, 177–179. [Google Scholar] [CrossRef] [Green Version]
- Almutairi, S.; Edrada-Ebel, R.; Fearnley, J.; Igoli, J.O.; Alotaibi, W.; Clements, C.J.; Gray, A.I.; Watson, D.G. Isolation of diterpenes and flavonoids from a new type of propolis from Saudi Arabia. Phytochem. Lett. 2014, 10, 160–163. [Google Scholar] [CrossRef]
- Alqarni, A.S.; Rushdi, A.I.; Owayss, A.A.; Raweh, H.S.; El-Mubarak, A.H.; Simoneit, B.R.T. Organic tracers from asphalt in propolis produced by urban honey bees, Apis mellifera Linn. PLoS ONE 2015, 10, e0128311. [Google Scholar] [CrossRef] [PubMed]
- Elnakady, Y.A.; Rushdi, A.I.; Franke, R.; Abutaha, N.; Ebaid, H.; Baabbad, M.; Omar, M.O.M.; Al Ghamdi, A.A. Characteristics, chemical compositions and biological activities of propolis from Al-Bahah, Saudi Arabia. Sci. Rep. 2017, 7, 41453. [Google Scholar] [CrossRef] [Green Version]
- Jerz, G.; Elnakady, Y.A.; Braun, A.; Jäckel, K.; Sasse, F.; Al Ghamdi, A.A.; Omar, M.O.M.; Winterhalter, P. Preparative mass-spectrometry profiling of bioactive metabolites in Saudi-Arabian propolis fractionated by high-speed countercurrent chromatography and off-line atmospheric pressure chemical ionization mass-spectrometry injection. J. Chromatogr. A 2014, 1347, 17–29. [Google Scholar] [CrossRef]
- Said, S.A.; Khan, S.A.; Ahmad, I.; Ali, H.S. Chemical composition of Egyptian and UAE propolis. Pak. J. Pharm. Sci. 2006, 19, 58–61. [Google Scholar]
- Narter, F.; Diren, A.; Kafkasli, A.; Eronat, A.P.; Seyhan, M.F.; Yilmaz-Aydogan, H.; Sarikaya, S.; Hatipoglu, S.D.; Sarica, K.; Ozturk, O. Anatolian propolis prevents oxalate kidney stones: Dramatic reduction of crystal deposition in ethylene-glycol-induced rat model. Rec. Nat. Prod. 2018, 12, 445–459. [Google Scholar] [CrossRef]
- Seyhan, M.F.; Yılmaz, E.; Timirci-Kahraman, Ö.; Saygılı, N.; Kısakesen, H.İ.; Gazioğlu, S.; Gören, A.C.; Eronat, A.P.; Begüm Ceviz, A.; Öztürk, T.; et al. Different propolis samples, phenolic content, and breast cancer cell lines: Variable cytotoxicity ranging from ineffective to potent. IUBMB Life 2019, 71, 619–631. [Google Scholar] [CrossRef]
- Aru, B.; Güzelmeric, E.; Akgül, A.; Demirel, G.Y.; Kırmızıbekmez, H. Antiproliferative Activity of Chemically Characterized Propolis from Turkey and Its Mechanisms of Action. Chem. Biodivers. 2019, 16, e1900189. [Google Scholar] [CrossRef]
- Çelemli, Ö.G.; Hatjina, F.; Charistos, L.; Schiesser, A.; Özkırım, A. More insight into the chemical composition of Greek propolis; Differences and similarities with Turkish propolis. Z. Nat. C 2013, 68, 429–438. [Google Scholar]
- Tugba Degirmencioglu, H.; Guzelmeric, E.; Yuksel, P.I.; Kırmızıbekmez, H.; Deniz, I.; Yesilada, E. A New Type of Anatolian Propolis: Evaluation of Its Chemical Composition, Activity Profile and Botanical Origin. Chem. Biodivers. 2019, 16, e1900492. [Google Scholar] [CrossRef] [PubMed]
- Duran, N.; Muz, M.; Culha, G.; Duran, G.; Ozer, B. GC-MS analysis and antileishmanial activities of two Turkish propolis types. Parasitol. Res. 2011, 108, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Gülçin, I.; Bursal, E.; Şehitoĝlu, M.H.; Bilsel, M.; Gören, A.C. Polyphenol contents and antioxidant activity of lyophilized aqueous extract of propolis from Erzurum, Turkey. Food Chem. Toxicol. 2010, 48, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Guzelmeric, E.; Ristivojević, P.; Trifković, J.; Dastan, T.; Yilmaz, O.; Cengiz, O.; Yesilada, E. Authentication of Turkish propolis through HPTLC fingerprints combined with multivariate analysis and palynological data and their comparative antioxidant activity. LWT Food Sci. Technol. 2018, 87, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Kargar, N.; Matin, G.; Matin, A.A.; Buyukisik, H.B. Biomonitoring, status and source risk assessment of polycyclic aromatic hydrocarbons (PAHs) using honeybees, pine tree leaves, and propolis. Chemosphere 2017, 186, 140–150. [Google Scholar] [CrossRef]
- Kartal, M.; Kaya, S.; Kurucu, S. GC-MS analysis of propolis samples from two different regions of Turkey. Z. Naturforsch. Sect. C J. Biosci. 2002, 57, 905–909. [Google Scholar] [CrossRef] [Green Version]
- Keskin, N.; Hazir, S.; Baser, K.H.C.; Kürkçüoglu, M. Antibacterial activity and chemical composition of Turkish propolis. Z. Naturforsch. C 2001, 56, 1112–1115. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Liao, L.; Wang, B. Potential antinociceptive effects of Chinese propolis and identification on its active compounds. J. Immunol. Res. 2018, 2018, 1–6. [Google Scholar] [CrossRef]
- Ahn, M.R.; Kumazawa, S.; Usui, Y.; Nakamura, J.; Matsuka, M.; Zhu, F.; Nakayama, T. Antioxidant activity and constituents of propolis collected in various areas of China. Food Chem. 2007, 101, 1383–1392. [Google Scholar] [CrossRef]
- Lee, I.-K.; Han, M.-S.; Kim, D.-W.; Yun, B.-S. Phenylpropanoid acid esters from Korean propolis and their antioxidant activities. Bioorg. Med. Chem. Lett. 2014, 24, 3503–3505. [Google Scholar] [CrossRef]
- Shimomura, K.; Sugiyama, Y.; Nakamura, J.; Ahn, M.R.; Kumazawa, S. Component analysis of propolis collected on Jeju Island, Korea. Phytochemistry 2013, 93, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Jin, U.H.; Chung, T.W.; Kang, S.K.; Suh, S.J.; Kim, J.K.; Chung, K.H.; Gu, Y.H.; Suzuki, I.; Kim, C.H. Caffeic acid phenyl ester in propolis is a strong inhibitor of matrix metalloproteinase-9 and invasion inhibitor: Isolation and identification. Clin. Chim. Acta 2005, 362, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, K.; Inui, S.; Sugiyama, Y.; Kurosawa, M.; Nakamura, J.; Choi, S.J.; Ahn, M.R.; Kumazawa, S. Identification of the plant origin of propolis from jeju Island, Korea, by observation of honeybee behavior and phytochemical analysis. Biosci. Biotechnol. Biochem. 2012, 76, 2135–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, M.; Chen, C.; Chen, P.; Huang, C.; Bankova, V. A validated spectrophotometric method for quantification of prenylated flavanones in pacific propolis from Taiwan. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2010, 21, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Ye, S.-R.; Ting, C.; Yu, Y.-H. Antibacterial activity of propolins from Taiwanese green propolis. J. Food Drug Anal. 2018, 26, 761–768. [Google Scholar] [CrossRef] [Green Version]
- Athikomkulchai, S.; Awale, S.; Ruangrungsi, N.; Ruchirawat, S.; Kadota, S. Chemical constituents of Thai propolis. Fitoterapia 2013, 88, 96–100. [Google Scholar] [CrossRef]
- Boonsai, P.; Phuwapraisirisan, P.; Chanchao, C. Antibacterial activity of a cardanol from Thai Apis mellifera propolis. Int. J. Med. Sci. 2014, 11, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Sanpa, S.; Popova, M.; Bankova, V.; Tunkasiri, T.; Eitssayeam, S.; Chantawannakul, P. Antibacterial compounds from propolis of Tetragonula laeviceps and Tetrigona melanoleuca (Hymenoptera: Apidae) from Thailand. PLoS ONE 2015, 10, e0126886. [Google Scholar] [CrossRef]
- Sun, L.; Wang, K.; Xu, X.; Ge, M.; Chen, Y.; Hu, F. Potential Protective Effects of Bioactive Constituents from Chinese Propolis against Acute Oxidative Stress Induced by Hydrogen Peroxide in Cardiac H9c2 Cells. Evid.-Based Complement. Altern. Med. 2017, 2017, 7074147. [Google Scholar] [CrossRef]
- Teerasripreecha, D.; Phuwapraisirisan, P.; Puthong, S.; Kimura, K.; Okuyama, M.; Mori, H.; Kimura, A.; Chanchao, C. In vitro antiproliferative/cytotoxic activity on cancer cell lines of a cardanol and a cardol enriched from Thai Apis mellifera propolis. BMC Complement. Altern. Med. 2012, 12, 27. [Google Scholar] [CrossRef] [Green Version]
- Chewchinda, S.; Vongsak, B. Development and validation of a high-performance thin layer chromatography method for the simultaneous quantitation of α- and γ-mangostins in Thai stingless bee propolis. Rev. Bras. Farmacogn. 2019, 29, 333–338. [Google Scholar] [CrossRef]
- Kumazawa, S.; Hamasaka, T.; Nakayama, T. Antioxidant activity of propolis of various geographic origins. Food Chem. 2004, 84, 329–339. [Google Scholar] [CrossRef]
- Georgieva, K.; Popova, M.; Dimitrova, L.; Trusheva, B.; Thanh, L.N.; Lan Phuong, D.T.; Lien, N.T.P.; Najdenski, H.; Bankova, V. Phytochemical analysis of Vietnamese propolis produced by the stingless bee Lisotrigona cacciae. PLoS ONE 2019, 14, e0216074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, M.; Trusheva, B.; Bankova, V. Propolis of stingless bees: A phytochemist’s guide through the jungle of tropical biodiversity. Phytomedicine 2021, 86, 153098. [Google Scholar] [CrossRef]
- Nguyen, H.X.; Nguyen, M.T.T.; Nguyen, N.T.; Awale, S. Chemical Constituents of Propolis from Vietnamese Trigona minor and Their Antiausterity Activity against the PANC-1 Human Pancreatic Cancer Cell Line. J. Nat. Prod. 2017, 80, 2345–2352. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.A.; Bayaqoob, N.I.M.; Rushdi, A.I.; Alattal, Y.; Simoneit, B.R.T.; El-Mubarak, A.H.; Al-Mutlaq, K.F. Chemical compositions and characteristics of organic compounds in propolis from Yemen. Saudi J. Biol. Sci. 2017, 24, 1094–1103. [Google Scholar] [CrossRef]
- Ozdal, T.; Ceylan, F.D.; Eroglu, N.; Kaplan, M.; Olgun, E.O.; Capanoglu, E. Investigation of antioxidant capacity, bioaccessibility and LC-MS/MS phenolic profile of Turkish propolis. Food Res. Int. 2019, 122, 528–536. [Google Scholar] [CrossRef]
- Popova, M.; Silici, S.; Kaftanoglu, O.; Bankova, V. Antibacterial activity of Turkish propolis and its qualitative and quantitative chemical composition. Phytomedicine 2005, 12, 221–228. [Google Scholar] [CrossRef]
- Silici, S.; Kutluca, S. Chemical composition and antibacterial activity of propolis collected by three different races of honeybees in the same region. J. Ethnopharmacol. 2005, 99, 69–73. [Google Scholar] [CrossRef]
- Chang, H.; Yuan, W.; Wu, H.; Yin, X.; Xuan, H. Bioactive components and mechanisms of Chinese poplar propolis alleviates oxidized low-density lipoprotein-induced endothelial cells injury. BMC Complement. Altern. Med. 2018, 18, 142. [Google Scholar] [CrossRef] [Green Version]
- Silici, S.; Ünlü, M.; Vardar-Ünlü, G. Antibacterial activity and phytochemical evidence for the plant origin of Turkish propolis from different regions. World J. Microbiol. Biotechnol. 2007, 23, 1797–1803. [Google Scholar] [CrossRef] [PubMed]
- Sorkun, K.; Süer, B.; Salih, B. Determination of chemical composition of Turkish propolis. Z. Naturforsch. C 2001, 56, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Velikova, M.; Bankova, V.; Sorkun, K.; Houcine, S.; Tsvetkova, I.; Kujumgiev, A. Propolis from the Mediterranean region: Chemical composition and antimicrobial activity. Z. Naturforsch. C 2000, 55, 790–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koru, O.; Toksoy, F.; Acikel, C.H.; Tunca, Y.M.; Baysallar, M.; Uskudar Guclu, A.; Akca, E.; Ozkok Tuylu, A.; Sorkun, K.; Tanyuksel, M.; et al. In vitro antimicrobial activity of propolis samples from different geographical origins against certain oral pathogens. Anaerobe 2007, 13, 140–145. [Google Scholar] [CrossRef]
- Hanif, A.; Nurdiansyah, R.; Hawa, P.; Wahyu, D.; Eko, K.; Dwi, C.; Taufiqu, N.; Novan, N.; Noviyanto, A.; Mardliyati, E. In silico investigation of potential inhibitors to main protease and spike protein of SARS-CoV-2 in propolis. Biochem. Biophys. Rep. 2021, 26, 100969. [Google Scholar] [CrossRef]
- Nakamura, R.; Nakamura, R.; Watanabe, K.; Oka, K.; Ohta, S.; Mishima, S.; Teshima, R. Effects of propolis from different areas on mast cell degranulation and identification of the effective components in propolis. Int. Immunopharmacol. 2010, 10, 1107–1112. [Google Scholar] [CrossRef]
- Sun, C.; Wu, Z.; Wang, Z.; Zhang, H. Effect of ethanol/water solvents on phenolic profiles and antioxidant properties of Beijing propolis extracts. Evid. Based Complement. Altern. Med. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Aronoff, S.L.; Berkowitz, K.; Shreiner, B.; Want, L. Glucose Metabolism and Regulation: Beyond Insulin and Glucagon. Diabetes Spectr. 2004, 17, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Meduru, H.; Wang, Y.-T.; Tsai, J.J.P.; Chen, Y.-C. Finding a potential dipeptidyl peptidase-4 (DPP-4) inhibitor for type-2 diabetes treatment based on molecular docking, pharmacophore generation, and molecular dynamics simulation. Int. J. Mol. Sci. 2016, 17, 920. [Google Scholar] [CrossRef] [Green Version]
- Wagner, R.; Kaiser, G.; Gerst, F.; Christiansen, E.; Due-Hansen, M.E.; Grundmann, M.; Machicao, F.; Peter, A.; Kostenis, E.; Ulven, T.; et al. Reevaluation of fatty acid receptor 1 as a drug target for the stimulation of insulin secretion in humans. Diabetes 2013, 62, 2106–2111. [Google Scholar] [CrossRef] [Green Version]
- Kristinsson, H.; Bergsten, P.; Sargsyan, E. Free fatty acid receptor 1 (FFAR1/GPR40) signaling affects insulin secretion by enhancing mitochondrial respiration during palmitate exposure. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 3248–3257. [Google Scholar] [CrossRef] [PubMed]
- Seckl, J.R. 11β-hydroxysteroid dehydrogenases: Changing glucocorticoid action. Curr. Opin. Pharmacol. 2004, 4, 597–602. [Google Scholar] [CrossRef]
- Morton, N.M. Obesity and corticosteroids: 11β-Hydroxysteroid type 1 as a cause and therapeutic target in metabolic disease. Mol. Cell. Endocrinol. 2010, 316, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- González-Rodríguez, Á.; Más-Gutierrez, J.A.; Mirasierra, M.; Fernandez-Pérez, A.; Lee, Y.J.; Ko, H.J.; Kim, J.K.; Romanos, E.; Carrascosa, J.M.; Ros, M. Essential role of protein tyrosine phosphatase 1B in obesity-induced inflammation and peripheral insulin resistance during aging. Aging Cell 2012, 11, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Graham, T.E.; Yang, Q.; Blüher, M.; Hammarstedt, A.; Ciaraldi, T.P.; Henry, R.R.; Wason, C.J.; Oberbach, A.; Jansson, P.-A.; Smith, U.; et al. Retinol-Binding Protein 4 and Insulin Resistance in Lean, Obese, and Diabetic Subjects. N. Engl. J. Med. 2006, 354, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Ramana, K.V.; Chandra, D.; Srivastava, S.; Bhatnagar, A. Regulation of aldose reductase and the polyol pathway activity by nitric oxide. Chem. Biol. Interact. 2003, 143–144, 333–340. [Google Scholar] [CrossRef]
- Zhan, J.-Y.; Ma, K.; Zheng, Q.-C.; Yang, G.-H.; Zhang, H.-X. Exploring the interactional details between aldose reductase (AKR1B1) and 3-Mercapto-5H-1,2,4-triazino [5,6-b]indole-5-acetic acid through molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 37, 1724–1735. [Google Scholar] [CrossRef]
- Mahmood, N. A review of α-amylase inhibitors on weight loss and glycemic control in pathological state such as obesity and diabetes. Comp. Clin. Path. 2016, 25, 1253–1264. [Google Scholar] [CrossRef]
- Visinoni, S.; Khalid, N.F.I.; Joannides, C.N.; Shulkes, A.; Yim, M.; Whitehead, J.; Tiganis, T.; Lamont, B.J.; Favaloro, J.M.; Proietto, J.; et al. The role of liver fructose-1,6-bisphosphatase in regulating appetite and adiposity. Diabetes 2012, 61, 1122–1132. [Google Scholar] [CrossRef] [Green Version]
- Matschinsky, F.M.; Wilson, D.F. The central role of glucokinase in glucose homeostasis: A perspective 50 years after demonstrating the presence of the enzyme in islets of Langerhans. Front. Physiol. 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brás, N.F.; Santos-Martins, D.; Fernandes, P.A.; Ramos, M.J. Mechanistic Pathway on Human α-Glucosidase Maltase-Glucoamylase Unveiled by QM/MM Calculations. J. Phys. Chem. B 2018, 122, 3889–3899. [Google Scholar] [CrossRef] [PubMed]
- Burwinkel, B.; Bakker, H.D.; Herschkovitz, E.; Moses, S.W.; Shin, Y.S.; Kilimann, M.W. Mutations in the liver glycogen phosphorylase gene (PYGL) underlying glycogenosis type VI (Hers disease). Am. J. Hum. Genet. 1998, 62, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Mellado-Gil, J.M.; Cobo-Vuilleumier, N.; Gauthier, B.R. Islet β -Cell Mass Preservation and Regeneration in Diabetes Mellitus: Four Factors with Potential Therapeutic Interest. J. Transplant. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monsalve, F.A.; Pyarasani, R.D.; Delgado-Lopez, F.; Moore-Carrasco, R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediat. Inflamm. 2013, 2013, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.-C.; Liu, Y.-C.; Lee, C.-C.; Chen, C.Y.-C. Potential retinoid X receptor agonists for treating Alzheimer’s disease from traditional Chinese medicine. Evid. Based Complement. Altern. Med. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Chen, B.-W.; Li, W.-X.; Wang, G.-H.; Li, G.-H.; Liu, J.-Q.; Zheng, J.-J.; Wang, Q.; Li, H.-J.; Dai, S.-X.; Huang, J.-F. A strategy to find novel candidate anti-Alzheimer’s disease drugs by constructing interaction networks between drug targets and natural compounds in medical plants. PeerJ 2018, 6, e4756. [Google Scholar] [CrossRef] [Green Version]
- Zander, M.; Madsbad, S.; Madsen, J.L.; Holst, J.J. Effect of 6-week course of glucagon-like peptide 1 on glycaemic control, insulin sensitivity, and β-cell function in type 2 diabetes: A parallel-group study. Lancet 2002, 359, 824–830. [Google Scholar] [CrossRef]
- Pujirahayu, N.; Bhattacharjya, D.K.; Suzuki, T.; Katayama, T. α-Glucosidase Inhibitory Activity of Cycloartane-Type Triterpenes Isolated from Indonesian Stingless Bee Propolis and Their Structure–Activity Relationship. Pharmaceuticals 2019, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Pu, R.; Li, Y.; Wu, Z.; Li, C.; Miao, X.; Yang, W. Chemical compositions of propolis from China and the United States and their antimicrobial activities against penicillium notatum. Molecules 2019, 24, 3576. [Google Scholar] [CrossRef] [Green Version]
- Gourgari, E.; Wilhelm, E.E.; Hassanzadeh, H.; Aroda, V.R.; Shoulson, I. A comprehensive review of the FDA-approved labels of diabetes drugs: Indications, safety, and emerging cardiovascular safety data. J. Diabetes Complicat. 2017, 31, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Triplitt, C.L.; Abdul-Ghani, M.; Cersosimo, E. Novel agents for the treatment of type 2 diabetes. Diabetes Spectr. 2014, 27, 100–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asgharpour, F.; Moghadamnia, A.A.; Zabihi, E.; Kazemi, S.; Ebrahimzadeh Namvar, A.; Gholinia, H.; Motallebnejad, M.; Nouri, H.R. Iranian propolis efficiently inhibits growth of oral streptococci and cancer cell lines. BMC Complement. Altern. Med. 2019, 19, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, H.; He, L. Current understanding of metformin effect on the control of hyperglycemia in diabetes. J. Endocrinol. 2016, 228, R97–R106. [Google Scholar] [CrossRef] [Green Version]
- Rena, G.; Hardie, D.G.; Pearson, E.R. The mechanisms of action of metformin. Diabetologia 2017, 60, 1577–1585. [Google Scholar] [CrossRef] [Green Version]
- Iheagwam, F.N.; Ogunlana, O.O.; Chinedu, S.N. Model optimization and in silico analysis of potential dipeptidyl peptidase IV antagonists from GC-MS identified compounds in Nauclea latifolia leaf extracts. Int. J. Mol. Sci. 2019, 20, 5913. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Lei, T.; Li, Y.; Song, Y.; Li, D.; Sun, H.; Hou, T. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J. Cheminform. 2016, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Lin, F.; Hasegawa, M.; Nojiri, H.; Yamane, H.; Okada, K. The potential bioproduction of the pharmaceutical agent sakuranetin, a flavonoid phytoalexin in rice. Bioengineered 2012, 3, 352–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerns, E.H.; Di, L.; Properties, D. Concepts, Structure Design and Methods: From ADME to Toxicity Optimization; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Saito, T.; Abe, D.; Sekiya, K. Sakuranetin induces adipogenesis of 3T3-L1 cells through enhanced expression of PPARγ2. Biochem. Biophys. Res. Commun. 2008, 372, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Y.; Sun, Y.; Zhang, G.; Bai, J.; Guo, J.; Su, X.; Du, H.; Cao, X. Yang Jet al. Naringenin Improves Insulin Sensitivity in Gestational Diabetes Mellitus Mice through AMPK. Nutr. Diabetes 2019, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liang, X.; Zhang, G.; Kong, L.; Peng, W.; Zhang, H. Galangin and Pinocembrin from Propolis Ameliorate Insulin Resistance in HepG2 Cells via Regulating Akt/mTOR Signaling. Evid. Based Complement. Altern. Med. 2018, 2018, 7971842. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Imm, J.-Y. The effect of chrysin-loaded phytosomes on insulin resistance and blood sugar control in type 2 diabetic db/db mice. Molecules 2020, 25, 5503. [Google Scholar] [CrossRef]
- Nagao, T.; Meguro, S.; Hase, T.; Otsuka, K.; Komikado, M.; Tokimitsu, I.; Yamamoto, T.; Yamamoto, K. A catechin-rich beverage improves obesity and blood glucose control in patients with type 2 diabetes. Obesity 2009, 17, 310–317. [Google Scholar] [CrossRef]
- Yang, Q.; Graham, T.E.; Mody, N.; Preitner, F.; Peroni, O.D.; Zabolotny, J.M.; Kotani, K.; Quadro, L.; Kahn, B.B. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005, 436, 356–362. [Google Scholar] [CrossRef]
- Sever, B.; Altıntop, M.D.; Demir, Y.; Akalın Çiftçi, G.; Beydemir, Ş.; Özdemir, A. Design, synthesis, in vitro and in silico investigation of aldose reductase inhibitory effects of new thiazole-based compounds. Bioorg. Chem. 2020, 102, 104110. [Google Scholar] [CrossRef]
- Martínez, L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef] [Green Version]
- Abdalla, M.; Eltayb, W.A.; El-Arabey, A.A.; Singh, K.; Jiang, X. Molecular dynamic study of SARS-CoV-2 with various S protein mutations and their effect on thermodynamic properties. Comput. Biol. Med. 2022, 141, 105025. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O. V Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Likić, V.A.; Gooley, P.R.; Speed, T.P.; Strehler, E.E. A statistical approach to the interpretation of molecular dynamics simulations of calmodulin equilibrium dynamics. Protein Sci. 2005, 14, 2955–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirolli, D.; Sciandra, F.; Bozzi, M.; Giardina, B.; Brancaccio, A.; De Rosa, M.C. Insights from molecular dynamics simulations: Structural basis for the V567D mutation-induced instability of zebrafish alpha-dystroglycan and comparison with the murine model. PLoS ONE 2014, 9, e103866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahamad, S.; Kanipakam, H.; Birla, S.; Ali, M.S.; Gupta, D. Screening Malaria-box compounds to identify potential inhibitors against SARS-CoV-2 Mpro, using molecular docking and dynamics simulation studies. Eur. J. Pharmacol. 2021, 890, 173664. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Patlewicz, G.; Jeliazkova, N.; Safford, R.J.; Worth, A.P.; Aleksiev, B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR QSAR Environ. Res. 2008, 19, 495–524. [Google Scholar] [CrossRef]
- BIOVIA. Discovery Studio Visualizer; Dassault Systems: San Diego, CA, USA, 2017. [Google Scholar]
- James, M.; Murtola, T.; Schulz, R.; Smith, J.C.; Hess, B.; Lindahl, E. ScienceDirect GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]






| No. | Drug Name | Total Similar Compounds | Potential Compound with the Highest Similarity (Percentage of Similarity) | T2DM Target (Docking Score, kcal/mol) |
|---|---|---|---|---|
| 1 | Chlorpropamide | 9 | 3′-Deoxysappanol (85.17%) | GCK (−9.4), AKR1B1 (−9.4) |
| 2 | Metformin | 2 | 1,4-Dihydrophenanthrene (84%) | AKR1B1 (−10.3) |
| 3 | Miglitol | 6 | Calamenene (85.35%) | RBP4 (−9.0) |
| 4 | Pioglitazone | 1 | TB-5 (81.2%) | FFAR1 (−9.6), HSD11B1 (−9.4), DPP4 (−9.4), PPARD (−9.3), GCK (−10.0), AKR1B1 (−11.3), PPARG (−9.4), RXRA (−9.9) |
| 5 | Nateglinide | 9 | Neobavaisoflavone (84.4%) | FFAR1 (−9.8), HSDB11B1 (−9.9), PPARD (−9.3), GCK (−9.9), RBP4 (−10.4), PPARG (−9.4), PPARG (−9.1) |
| 6 | Tolazamide | 11 | 4-hydroxy-1-(2-hydroxyphenyl)-4-phenylbut-2-en-1-one (85.17%) | FFAR1 (−9.9), AKR1B1 (−9.5) |
| 7 | Tolbutamide | 10 | 6-hydroxy-3-methoxy-6-(3-phenyl-2-propenyl)-2-cyclehexane-1-one (84.03%) | FFAR1 (−9.2), AKR1B1 (−9.4) |
| 8 | Rosiglitazone | 3 | Phenethyl ferulate (81.5%) | FFAR1 (−9.3), AKR1B1 (−9.8), RXRA (−9.3) |
| 9 | Gliclazide | 17 | Benzyl caffeate (86.5%) | FFAR1 (−9.5), AKR1B1 (−9.8), RBP4 (−9.6), RXRA (−9.0) |
| 10 | Salsalate | 40 | 1-phenanthrenecarboxylic acid (87.84%) | AKR1B1 (−10.1), RBP4 (−9.8) |
| 11 | Lipoic acid | 18 | trans-chalcone (90.41%) | FFAR1 (−9.7), AKR1B1 (−9.4) |
| 12 | Vildagliptin | 3 | Tschimganine (84.18%) | AKR1B1 (−9.3), RBP4 (−9.6) |
| 13 | Glymidine | 3 | Phenethyl caffeate (81.12%) | FFAR1 (−9.4), GCK (−9.2), AKR1B1 (−10.3), RBP4 (−9.8), RXRA (−9.1) |
| 14 | Ipragliflozin | 1 | Phenethyl ferulate (81%) | FFAR1 (−9.3), AKR1B1 (−9.8), RXRA (−9.3) |
| 15 | Voglibose | 5 | Delta-cadinene (84.54%) | RBP4 (−9.0) |
| 16 | Omarigliptin | 2 | Rosmarinic acid (81%) | AKR1B1 (−10.5), PPARG (−9.1) |
| 17 | Serotonin | 12 | 1,4-Dihydrophenanthrene (87.9%) | AKR1B1 (−10.3) |
| 18 | Mitiglinide | 10 | Ferulic acid benzyl ester (83.4%) | FFAR1 (−9.5), AKR1B1 (−9.7) |
| ADMET Property | Potential Compounds (%) | Ratio of Potential Compounds | Acceptable Range/Criteria |
|---|---|---|---|
| Lipinski’s rules | 91.6 | 252/275 | 0–1 violation |
| Veber’s rules | 92.4 | 254/275 | 0 violation |
| Solubility (Log S) | 45.4 | 124/275 | >10 µg/mL |
| Caco-2 permeability | 74.5 | 205/275 | >−5.15 log unit |
| Protein plasma binding | 31.6 | 87/275 | >90% |
| Blood–brain barrier (BBB) | 73.1 | 201/275 | Category 1: BBB+ |
| Human intestinal absorption (HIA) | 95.6 | 263/275 | >30%; Category 1 HIA+ |
| LD50 of acute toxicity | 60.0 | 165/275 | >500 mg/kg |
| Human hepatotoxicity (H-HT) | 54.2 | 126/275 | Category 0: H-HT negative |
| Ames Mutagenicity | 80.7 | 222/275 | Category 0: Ames negative |
| Potential carcinogen | 100 | 275/275 | No |
| hERG blockers | 73.8 | 202/275 | Category 0: Non-blockers |
| Potential Compounds | Lipinski’s Rules (Violations) | Veber’s Rules (Violations) | MW (g/mol) | XLOGP3 | H-Bond Acceptors | H-Bond Donors | Rotatable Bonds | TPSA (Å) | logS (µg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| (2R)-7,4′-Dihydroxy-5-methoxy-8-methylflavane | 0 | 0 | 286.32 | 3.44 | 4 | 2 | 2 | 58.92 | 16.36 |
| (RR)-(+)-3′-senecioylkhellactone | 0 | 0 | 218.33 | 3.94 | 1 | 1 | 1 | 20.23 | 35.65 |
| 2′,4′,6′-Trihydroxy chalcone (pinocembrin chalcone) | 0 | 0 | 256.25 | 3.18 | 4 | 3 | 3 | 77.76 | 82.54 |
| Alpinetin | 0 | 0 | 270.28 | 2.65 | 4 | 1 | 2 | 55.76 | 32.57 |
| Catechin | 0 | 0 | 290.27 | 0.36 | 6 | 5 | 1 | 110.38 | 273.40 |
| Chrysin | 0 | 0 | 254.24 | 3.52 | 4 | 2 | 1 | 70.67 | 49.69 |
| Hesperetin | 0 | 0 | 302.28 | 2.60 | 6 | 3 | 2 | 96.22 | 94.93 |
| Naringenin | 0 | 0 | 272.25 | 2.52 | 5 | 3 | 1 | 86.99 | 83.94 |
| Pinobanksin-3-O-butyrate | 0 | 0 | 342.34 | 3.76 | 6 | 2 | 5 | 93.06 | 34.87 |
| Pinocembrin | 0 | 0 | 256.25 | 2.88 | 4 | 2 | 1 | 66.76 | 51.25 |
| Pinocembrin-5-methyl ether | 0 | 0 | 270.28 | 2.65 | 4 | 1 | 2 | 55.76 | 32.57 |
| Sakuranetin | 0 | 0 | 286.28 | 2.85 | 5 | 2 | 2 | 75.99 | 58.18 |
| Potential Compounds | Caco-2 Permeability (Log Unit) | Human Intestinal Absorption | Blood–Brain Barrier | Protein Plasma Binding (PPB)% |
|---|---|---|---|---|
| (2R)-7,4′-Dihydroxy-5-methoxy-8-methylflavane | −4.854 | + | + | 90.46 |
| (RR)-(+)-3′-senecioylkhellactone | −4.335 | + | + | 85.05 |
| 2′,4′,6′-Trihydroxy chalcone (pinocembrin chalcone) | −4.904 | + | + | 88.97 |
| Alpinetin | −4.644 | + | + | 88.56 |
| Catechin | −6.250 | + | + | 93.86 |
| Chrysin | −4.973 | + | + | 89.57 |
| Hesperetin | −4.876 | + | + | 88.06 |
| Naringenin | −4.781 | + | + | 89.09 |
| Pinobanksin-3-O-butyrate | −4.987 | + | + | 89.10 |
| Pinocembrin | −4.882 | + | + | 87.44 |
| Pinocembrin-5-methyl ether | −4.644 | + | + | 88.56 |
| Sakuranetin | −4.830 | + | + | 89.56 |
| Potential Compounds | LD50 Acute Toxicity (mg/kg) | Hepatotoxicity | Ames Mutagenicity | hERG Blockers | Tumorigenic | Potential Carcinogen Based on QSAR |
|---|---|---|---|---|---|---|
| (2R)-7,4′-Dihydroxy-5-methoxy-8-methylflavane | 784.98 | No | No | No | No | No |
| (RR)-(+)-3′-senecioylkhellactone | 1535.08 | No | No | No | low | No |
| 2′,4′,6′-Trihydroxy chalcone (pinocembrin chalcone) | 1975.49 | No | No | No | No | No |
| Alpinetin | 1293.66 | No | No | No | No | No |
| Catechin | 860.6 | No | No | No | No | No |
| Chrysin | 1054.98 | No | No | No | No | No |
| Hesperetin | 857.85 | No | No | No | No | No |
| Naringenin | 995.35 | No | No | No | No | No |
| Pinobanksin-3-O-butyrate | 599.05 | No | No | No | No | No |
| Pinocembrin | 1311.22 | No | No | No | No | No |
| Pinocembrin-5-methyl ether | 1293.66 | No | No | No | No | No |
| Sakuranetin | 872.56 | No | No | No | No | No |
| Compound | Predicted Targets (Docking Score in kcal/mol) | Potential Antidiabetic Effect | Country of Origin |
|---|---|---|---|
(2R)-7,4′-Dihydroxy-5-methoxy-8-methylflavane | RBP4 (−9.7) | Regulation of insulin secretion and sensitivity | Vietnam |
(RR)-(+)-3′-senecioylkhellactone | HSD11B1 (−9.5), PPARD (−10.0), GCK (−9.4), AKR1B1 (−9.4), PPARG (−9.1), RXRA (−9.4) | Regulation of insulin secretion and sensitivity, regulation of glucose and lipid metabolism | South Korea |
2′,4′,6′-Trihydroxy chalcone (pinocembrin chalcone) | FFAR1 (−9.4), AKR1B1 (−9.0) | Regulation of insulin secretion/sensitivity and glucose metabolism | Iran |
Alpinetin | RBP4 (−9.3) | Regulation of insulin secretion and sensitivity | Beijing, China |
Catechin | AKR1B1 (−9.0),RBP4 (−9.0) | Regulation of insulin secretion/sensitivity and glucose metabolism | China |
Chrysin | AKR1B1 (−9.0), RBP4 (−9.6), RXRA (−9.2) | Regulation of insulin secretion/sensitivity and glucose metabolism | Jordan, China, Thailand, Turkey, Iraq, Indonesia, Iran, Lebanon, South Korea, Uzbekistan |
Hesperetin | FFAR1 (−9.0), HSD11B1 (−9.3), RBP4 (−9.6) | Regulation of insulin secretion/sensitivity and glucose metabolism | Iraq, China |
Naringenin | RBP4 (−9.7) | Regulation of insulin secretion/sensitivity | Jordan, Turkey, Iran, Iraq, China, |
Pinobanksin-3-O-butyrate | AKR1B1 (−9.0) | Regulation of glucose metabolism | China, Iran |
Pinocembrin | RBP4 (−9.6) | Regulation of insulin secretion/sensitivity | India, Lebanon, South Korea, Uzbekistan, Jordan, Iraq, Iran, China, Turkey, Nepal, Thailand |
Pinocembrin-5-methyl ether | RBP4 (−9.4) | Regulation of insulin secretion/sensitivity | China |
Sakuranetin | RBP4 (−9.6) | Regulation of insulin secretion/sensitivity | Iraq, Turkey, China |
| Potential Compounds (Number Code) | RBP4 | AKR1B1 |
|---|---|---|
| (2R)-7,4′-Dihydroxy-5-methoxy-8-methylflavane (21) | Ala55 (C-H-bond), Arg121 (π-Cation), Met88 (π-Sulfur, Alkyl), Tyr90 (π-π T-Shaped, π-Alkyl interaction), Ala57 (π-Alkyl, Alkyl interaction), Leu37 (π-Alkyl, Alkyl interaction) Total: 6 residues | (Binding score > −9.0 Kcal/mol) |
| (RR)-(+)-3′-senecioylkhellactone (41) | (Binding score > −9.0 Kcal/mol) | Phe122 (π-π Stacked), Tyr209 (π-Sigma, π-Alkyl) Total: 2 residues |
| 2′,4′,6′-Trihydroxy chalcone (pinocembrin chalcone) (91) | (Binding score > −9.0 Kcal/mol) | Cys303 (π-Sulfur), Cys80 (π-Sulfur), Leu300 (π-alkyl), Trp111 (π-π Stacked) Trp20 (π-π Stacked, π-π T-shaped), Cys298 (π-Sulfur) Total: 6 residues |
| Alpinetin (288) | Tyr133 (π-π Stacked), Arg121 (π-cation interaction), Pro32 (π-Alkyl) Leu37 (π-Alkyl), Met88 (π-sulfur), Ala57 (π-Alkyl, Alkyl interaction), Ala55 (Alkyl interaction) Total: 7 residues | (Binding score > −9.0 Kcal/mol) |
| Catechin (353) | Lys29 (H-bond), Pro32 (C-H bond), Tyr133 (π-π Stacked) Leu37 (π-Alkyl), Ala55 (π-Alkyl), Met88 (π-Sulfur) Ala57 (π-Alkyl) Total: 7 residues | Trp20 (π-π T-shaped), Cys80 (π-Sulfur), Leu300 (π-alkyl), Val47 (π-alkyl) Trp111 (π-π Stacked), Cys303 (π-alkyl) Total: 6 residues |
| Crysin (359) | Tyr90 (π-π T-shaped), Leu37 (π-Alkyl), Met73 (π-alkyl), Ala43 (π-alkyl), Ala57 (π-alkyl), Ala55 (π-alkyl), Met88 (π-sulfur) Total: 7 residues | Cys80 (π-sulfur), Cys303 (π-sulfur), Trp111 (H-Bond, π-π Stacked), Phe122 (π-π T-shaped), Leu300 (π-Alkyl) Total: 5 residues |
| Hesperetin (482) | Tyr90 (H-bond, π-π T-shaped), Arg121 (2 H-bond) Pro32 (Alkyl-interaction), Asp102 (π-Anion), Leu37 (π-Alkyl), Tyr133 (π-π Stacked), Ala43 (π-Alkyl) Ala57 (π-Alkyl), Ala 55 (H-bond, π-Alkyl) Total: 9 residues | (Binding score > −9.0 Kcal/mol) |
| Naringenin (568) | Arg121 (H-bond), Leu37 (π-Alkyl), Tyr90 (π-π T-shaped), Asp102 (π-anion) Thr56 (C-H Bond), Met88 (π-sulfur), Ala43 (π-alkyl), Ala55 (π-alkyl), Ala57 (π-alkyl) Total: 9 residues | (Binding score > −9.0 Kcal/mol) |
| Pinobanksin-3-O-butyrate (633) | (Binding score > −9.0 Kcal/mol) | Phe122 (π-π stacked), Val47 (H-bond, π-Alkyl), Trp20 (C-H bond, π-π stacked, π-Alkyl), Cys298 (H-bond), Tyr209 (π-sigma), Trp111 (H-bond, π-π stacked), Leu300 (π-alkyl) Total: 7 residues |
| Pinocembrin (640) | Arg121 (π-cation), Leu37 (π-alkyl), Tyr90 (π-π T-Shaped), Ala55 (π-alkyl), Ala43 (π-alkyl), Ala57 (π-alkyl), Met88 (π-sulfur) Total: 7 residues | (Binding score > −9.0 Kcal/mol) |
| Pinocembrin-5-methyl ether (641) | Pro32 (π-alkyl), Leu37 (π-alkyl), Arg121 (π-cation), Met88 (π-sulfur), His104 (C-H bond), Ala57 (π-alkyl, alkyl interaction), Ala55 (C-H bond), Tyr133 (π-π stacked) Total: 7 residues | (Binding score > −9.0 Kcal/mol) |
| Sakuranetin (681) | Lys29 (H-bond), Phe137 (π-alkyl), Ala57 (π-alkyl), Phe45 (π-alkyl), Ala43 (Alkyl interaction), Arg121 (π-cation), Leu37 (π-alkyl), His104 (π-alkyl), Met88 (π-sulfur) Total: 9 residues | (Binding score > −9.0 Kcal/mol) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syaifie, P.H.; Harisna, A.H.; Nasution, M.A.F.; Arda, A.G.; Nugroho, D.W.; Jauhar, M.M.; Mardliyati, E.; Maulana, N.N.; Rochman, N.T.; Noviyanto, A.; et al. Computational Study of Asian Propolis Compounds as Potential Anti-Type 2 Diabetes Mellitus Agents by Using Inverse Virtual Screening with the DIA-DB Web Server, Tanimoto Similarity Analysis, and Molecular Dynamic Simulation. Molecules 2022, 27, 3972. https://doi.org/10.3390/molecules27133972
Syaifie PH, Harisna AH, Nasution MAF, Arda AG, Nugroho DW, Jauhar MM, Mardliyati E, Maulana NN, Rochman NT, Noviyanto A, et al. Computational Study of Asian Propolis Compounds as Potential Anti-Type 2 Diabetes Mellitus Agents by Using Inverse Virtual Screening with the DIA-DB Web Server, Tanimoto Similarity Analysis, and Molecular Dynamic Simulation. Molecules. 2022; 27(13):3972. https://doi.org/10.3390/molecules27133972
Chicago/Turabian StyleSyaifie, Putri Hawa, Azza Hanif Harisna, Mochammad Arfin Fardiansyah Nasution, Adzani Gaisani Arda, Dwi Wahyu Nugroho, Muhammad Miftah Jauhar, Etik Mardliyati, Nurwenda Novan Maulana, Nurul Taufiqu Rochman, Alfian Noviyanto, and et al. 2022. "Computational Study of Asian Propolis Compounds as Potential Anti-Type 2 Diabetes Mellitus Agents by Using Inverse Virtual Screening with the DIA-DB Web Server, Tanimoto Similarity Analysis, and Molecular Dynamic Simulation" Molecules 27, no. 13: 3972. https://doi.org/10.3390/molecules27133972