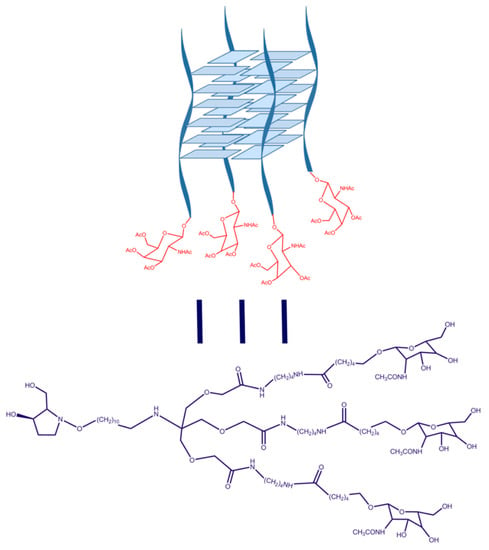
Properties of Parallel Tetramolecular G-Quadruplex Carrying N-Acetylgalactosamine as Potential Enhancer for Oligonucleotide Delivery to Hepatocytes
Abstract
:1. Introduction
2. Results
2.1. Oligonucleotides
2.2. Characterization of G-Quadruplex Formation
2.2.1. Characterization of G-Quadruplex Formation by CD
2.2.2. Characterization of G-Quadruplex Formation by NMR
2.2.3. Characterization of G-Quadruplex Formation by Gel Electrophoresis
2.3. ASGPR Receptor Binding Studies
2.4. Toxicity Assays
2.5. Analysis of the Stabilityof Oligonucleotides towards Snake Venom Phosphodiesterase and 10% FBS
2.6. Internalization Assay
2.7. Antisense Studies
3. Discussion
4. Materials and Methods
4.1. Synthesis of GalNac Oligonucleotide Conjugates and Controls
4.2. G-Quadruplex Formation
4.3. Circular Dichroism
4.4. Nuclear Magnetic Resonance (NMR) Experiments
4.5. Polyacrylamide Electrophoresis Assays
4.6. Affinity Binding to ASGPR Receptors in Primary Mouse Hepatocytes
4.7. MTT Assays
4.8. Flow Cytometry
4.9. Luciferase Assays
4.10. Analysis of the Stability of Oligonucleotides towards Snake Venom Phosphodiesterase and 10% FBS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Khvorova, A.; Watts, J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotech. 2017, 35, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Jorge, A.F.; Grijalvo, S.; Fàbrega, C.; Aviñó, A.; Eritja, R. Advances in therapeutic oligonucleotide chemistry. In Nucleic Acids Chemistry. Modifications and Conjugates for Biomedicine and Nanotechnology; Eritja, R., Ed.; De Gruyter: Berlin, Germany, 2021; pp. 273–329. [Google Scholar]
- Al Shaer, D.; Al Musaimi, O.; Albericio, F.; de la Torre, B.G. 2019 FDA TIDES (peptides and oligonucleotides) harvest. Pharmaceuticals 2020, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Liang, X.H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Manoharan, M. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 2019, 52, 1036–1047. [Google Scholar] [CrossRef]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham, E.T., Jr.; Feinsod, M.; Guyer, D.R. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, G.P.; Lee, S.S. Therapeutic miRNA and siRNA moving from bench to clinic as next generation on medicine. Mol. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef]
- Mendell, J.T. Targeting a long noncoding RNA in breast cancer. N. Engl. J. Med. 2016, 374, 2287–2289. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef]
- Balwani, M.; Sardh, E.; Ventura, P.; Aguilera Peiró, P.; Rees, D.C.; Stölzel, U.; Bissell, D.M.; Bonkovsky, H.L.; Windyga, J.; Anderson, K.E.; et al. Phase 3 trial of RNAi therapeutic Givosiran for acute intermittent porphyria. N. Engl. J. Med. 2020, 382, 2289–2301. [Google Scholar] [CrossRef]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.S.; Chan, A.; Charisse, K.; Alam, R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S.; et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef] [PubMed]
- Rajeev, K.G.; Nair, J.K.; Jayaraman, M.; Charisse, K.; Taneja, N.; O’Shea, J.; Willoughby, J.L.S.; Yucius, K.; Nguyen, T.; Shulga-Morskaya, S.; et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 2015, 16, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Keiser, K.; Nair, J.K.; Charisse, K.; Manoharan, R.M.; Kretschmer, P.; Peng, C.G.; Kel’in, A.V.; Kandasamy, P.; Willoughby, J.L.S.; et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015, 10, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Janas, M.M.; Schlegel, M.K.; Harbison, C.E.; Yilmaz, V.O.; Jiang, Y.; Parmar, R.; Zlatev, I.; Castoreno, A.; Xu, H.; Shulga-Morskaya, S.; et al. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018, 9, 723. [Google Scholar] [CrossRef]
- Gatto, B.; Palimbo, M.; Sissi, C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Koutsoudakis, G.; Paris de León, A.; Herrera, C.; Dorner, M.; Pérez-Vilaró, G.; Lyonnais, S.; Grijalvo, S.; Eritja, R.; Meyerhans, A.; Mirambeau, G.; et al. Oligonucleotide-lipid conjugates forming G-quadruplex structures are potent and pangenotypic hepatitis C virus entry inhibitors in vitro and ex vivo. Antimicrob. Agents Chemother. 2017, 61, e02354-16. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biophys. Biochip. Acta 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biophys. Biochim. Acta 2017, 1861, 1429–1447. [Google Scholar] [CrossRef]
- Riccardi, C.; Fàbrega, C.; Grijalvo, S.; Vitiello, G.; D’Errico, G.; Eritja, R.; Montesarchio, D. AS1411-decorated niosomes as effective nanocarriers for Ru(III)-based drugs in anticancer strategies. J. Mat. Chem. B 2018, 6, 5368–5384. [Google Scholar] [CrossRef]
- Grijalvo, S.; Alagia, A.; Gargallo, R.; Eritja, R. Cellular uptake studies of antisense oligonucleotides using g-quadruplex-nanostructures: The effect of cationic residue in the biophysical and biological properties. RSC Adv. 2016, 6, 76099–76109. [Google Scholar] [CrossRef]
- Grijalvo, S.; Clua, A.; Eres, M.; Gargallo, R.; Eritja, R. Tuning G-quadruplex nanostructures with lipids. Towards designing hybrid scaffolds for oligonucleotide delivery. Int. J. Mol. Sci. 2021, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Clua, A.; Fàbrega, C.; García-Chica, J.; Grijalvo, S.; Eritja, R. Parallel G-quadruplex structures increase cellular uptake and cytotoxicity of 5-fluoro-2′-deoxyuridine oligomers in 5-fluorouracil resistant cells. Molecules 2021, 26, 1741. [Google Scholar] [CrossRef] [PubMed]
- Lyonnais, S.; Hounsou, C.; Teulade-Fichou, M.P.; Jeusset, J.; Le Cam, E.; Mirambeau, G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res. 2002, 30, 5276–5283. [Google Scholar]
- Lyonnais, S.; Gorelick, R.J.; Mergny, J.L.; Le Cam, E.; Mirambeau, G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003, 31, 5754–5763. [Google Scholar] [CrossRef] [PubMed]
- Lyonnais, S.; Grijalvo, S.; Alvarez-Fernández, C.; Fleta, E.; Martínez, J.; Meyerhans, A.; Sánchez-Palomino, S.; Mirambeau, G.; Eritja, R. Lipid-oligonucleotide conjugates forming G-quadruplex (lipoquads) as potent inhibitors of HIV entry. Proceedings 2017, 1, 670. [Google Scholar]
- Zhang, H.-Y.; Mao, J.; Zhou, D.; Xu, D.; Thonberg, H.; Liang, Z.; Wahlestedt, C. mRNA accessible site tagging (MAST): A novel high throughput method for selecting effective antisense oligonucleotides. Nucleic Acid Res. 2003, 31, e72. [Google Scholar] [CrossRef]
- Malgowska, M.; Gudanis, D.; Teubert, A.; Dominiak, G.; Gdaniec, Z. How to study G-quadruplex structures. BioTechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2012, 93, 381–390. [Google Scholar] [CrossRef]
- Mergny, J.L.; De Cian, A.; Ghelab, A.; Sacca, B.; Lacroix, L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005, 33, 81–94. [Google Scholar] [CrossRef]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR spectroscopy of G-quadruplex. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Lin, C.; Dickerhoff, J.; Yang, D. NMR studies of G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol. Biol. 2019, 2035, 157–176. [Google Scholar]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Gros, J.; Rosu, F.; Amrane, S.; De Cian, A.; Gabelica, V.; Lacroix, L.; Mergny, J.L. Guanines are a quartet’s best friend: Impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007, 35, 3064–3075. [Google Scholar] [CrossRef] [PubMed]
- Gros, J.; Aviñó, A.; Lopez de la Osa, J.; González, C.; Lacroix, L.; Pérez, A.; Orozco, M.; Eritja, R.; Mergny, J.L. 8-Aminoguanine accelerates tetramolecular G-quadruplex formation. Chem. Comm. 2008, 2926–2928. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Sleiman, H.F. DNA nanostructures: Current challenges and opportunities for cellular delivery. ACS Nano 2021, 15, 3631–3645. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Christensen, N.J.; Martos-Maldonado, M.C.; Midtgaard, S.R.; Ejlersen, M.; Thulstrup, P.W.; Sørensen, K.K.; Jensen, K.J.; Wengel, J. Folding topology of a short coiled-coil peptide structure templated by an oligonucleotide triplex. Chem. Eur. J. 2017, 23, 9297–9305. [Google Scholar] [CrossRef]
- Ghosh, P.S.; Hamilton, A.D. Noncovalent template-assisted mimicry of multiloop protein surfaces: Assembling discontinuous and functional domains. J. Am. Chem. Soc. 2012, 134, 13208–13211. [Google Scholar] [CrossRef]
- Musumeci, D.; Montesarchio, D. Synthesis of a cholesterol-HEG phosphoramidite derivative and its application to lipid-conjugates of the anti-HIV 5′TGGGAG3′ Hotoda’s sequence. Molecules 2012, 17, 12378–12392. [Google Scholar] [CrossRef]










| N | Name | Sequence (5′-3′) | Backbone | GalNAc |
|---|---|---|---|---|
| H1 | control | TTGGGGGGTACAGTGCA | PO | - |
| H2 | G-PO-HCV-L235 | TTGGGGGGTACAGTGCA-L235 | PO | L235 |
| H3 | A-PO-HCV-L235 | TTGAAAGGTACAGTGCA-L235 | PO | L235 |
| H4 | G-PO-HCV-L193 | TTGGGGGGTACAGTGCA-L193 | PO | L193 |
| H5 | A-PO-HCV-L193 | TTGAAAGGTACAGTGCA-L193 | PO | L193 |
| H6 | FAM-G-PO-HCV-L235 | FAM-TTGGGGGGTACAGTGCA-L235 | PO | L235 |
| H7 | FAM-G-PO-HCV-L193 | FAM-TTGGGGGGTACAGTGCA-L193 | PO | L193 |
| H8 | A-PS-HCV-L235 | TsTsGAAAGGTACAGsTsGsCsA-L235 | PO/PS | L235 |
| H9 | A-PS-HCV-L193 | TsTsGAAAGGTACAGsTsGsCsA-L193 | PO/PS | L193 |
| H10 | G-PS-HCV-L235 | TsTsGGGGGGTACAGsTsGsCsA-L235 | PO/PS | L235 |
| H11 | G-PS-HCV-L193 | TsTsGGGGGGTACAGsTsGsCsA-L193 | PO/PS | L193 |
| H12 | A-PO-HCV-L96 | TTGAAAGGTACAGTGCA-L96 | PO | L96 |
| N | Name | Sequence (5′-3′) | Backbone | GalNAc |
|---|---|---|---|---|
| L1 | ASO control | CsGsTsTsTsCsCsTsTsTsGsTsTsCsTsGsGsA | PS | - |
| L2 | Gapmer control | CsGsUsUsTsCsCsTsTsTsGsTsTsCsUsGsGsA | PS | - |
| L3 | Luc-G6-GalNAc | CsGsUsUsTsCsCsTsTsTsGsTsTsCsUsGsGsATGGGGGGT-L193 | PO,PS | L193 |
| L4 | G6-Luc-GalNAc | TGGGGGGTCsGsUsUsTsCsCsTsTsTsGsTsTsCsUsGsGsAs-L193 | PO,PS | L193 |
| L5 | Luc-T4-GalNAc | CsGsUsUsTsCsCsTsTsTsGsTsTsCsUsGsGsATTTT-L193 | PO,PS | L193 |
| L6 | Scr-Luc-G6-GalNAc | CsUsGsUsCsTsGsAsCsGsTsTsCsTsUsUsGsUsTGGGGGGT-L193 | PO,PS | L193 |
| L7 | FAM-Luc-G6-GalNAc | FAM-sCsUsGsUsTsCsCsTsTsTsGsTsTsCsUsGsGsAsTTGGGGGGT-L193 | PO,PS | L193 |
| L8 | T8 | TTTTTTTT | PO | - |
| N | Name | KI (nM) | Stdev |
|---|---|---|---|
| H4 | G-PO-HCV-L193 | 47.2 | 7.3 |
| H2 | G-PO-HCV-L235 | 39.9 | 3.2 |
| H11 | G-PS-HCV-L193 | 42.8 | 7.6 |
| H10 | G-PS-HCV-L235 | 31.3 | 7.2 |
| H12 | A-PO-HCV-L96 | 10.4 | 3.2 |
| H5 | A-PO-HCV-L193 | N/D 1 | N/D 1 |
| N | Motif | KI (nM) | Stdev |
|---|---|---|---|
| R1 | Sense-L96 | 8.8 | 2.2 |
| R2 | Sense-TG6T-L193 | 696.0 | 37.2 |
| R2 + R3 | Duplex-TG6T-L193 | N/D 1 | N/D 1 |
| L5 | Luc-T4-L193 | 300.7 | 23.9 |
| L3 | Luc-G6-L193 | 357.8 | 48.5 |
| L4 | G6-Luc-L193 | 121.1 | 17.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clua, A.; Grijalvo, S.; Erande, N.; Gupta, S.; Yucius, K.; Gargallo, R.; Mazzini, S.; Manoharan, M.; Eritja, R. Properties of Parallel Tetramolecular G-Quadruplex Carrying N-Acetylgalactosamine as Potential Enhancer for Oligonucleotide Delivery to Hepatocytes. Molecules 2022, 27, 3944. https://doi.org/10.3390/molecules27123944
Clua A, Grijalvo S, Erande N, Gupta S, Yucius K, Gargallo R, Mazzini S, Manoharan M, Eritja R. Properties of Parallel Tetramolecular G-Quadruplex Carrying N-Acetylgalactosamine as Potential Enhancer for Oligonucleotide Delivery to Hepatocytes. Molecules. 2022; 27(12):3944. https://doi.org/10.3390/molecules27123944
Chicago/Turabian StyleClua, Anna, Santiago Grijalvo, Namrata Erande, Swati Gupta, Kristina Yucius, Raimundo Gargallo, Stefania Mazzini, Muthiah Manoharan, and Ramon Eritja. 2022. "Properties of Parallel Tetramolecular G-Quadruplex Carrying N-Acetylgalactosamine as Potential Enhancer for Oligonucleotide Delivery to Hepatocytes" Molecules 27, no. 12: 3944. https://doi.org/10.3390/molecules27123944