Stability of ZIF-8 Nanoparticles in Most Common Cell Culture Media
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of ZIF-8 Nanoparticles
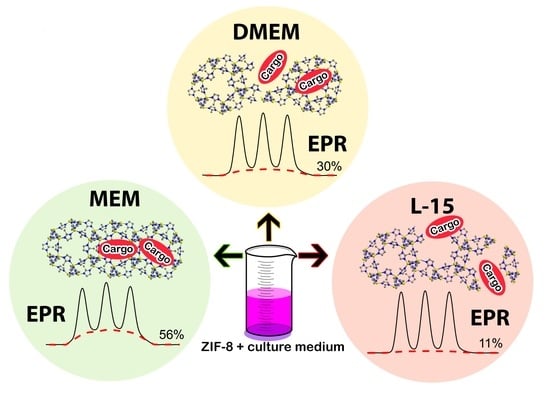
2.2. Dissolution of ZIF-8 in Cell Culture Media
2.3. Dissolution of ZIF-8 by Individual Amino Acids
2.4. Stabilization of ZIF-8 Nanoparticles by 2-Methylimidazole
3. Conclusions
4. Experimental
4.1. Materials
4.2. Synthesis of R
4.3. Synthesis of R@ZIF-8
4.4. Sample Preparation
4.5. Characterization of ZIF-8 Particles
4.6. EPR Measurements
4.7. Cytotoxic Assay of MIM
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, K.S.; Ni, Z.; Côté, A.P.; Choi, J.Y.; Huang, R.; Uribe-Romo, F.J.; Chae, H.K.; O’Keeffe, M.; Yaghi, O.M. Exceptional Chemical and Thermal Stability of Zeolitic Imidazolate Frameworks. Proc. Natl. Acad. Sci. USA 2006, 103, 10186–10191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, R.; Phan, A.; Wang, B.; Knobler, C.; Furukawa, H.; O’Keeffe, M.; Yaghi, O.M. High-Throughput Synthesis of Zeolitic Imidazolate Frameworks and Application to CO2 Capture. Science 2008, 319, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Deria, P.; Mondloch, J.E.; Karagiaridi, O.; Bury, W.; Hupp, J.T.; Farha, O.K. Beyond Post-Synthesis Modification: Evolution of Metal-Organic Frameworks via Building Block Replacement. Chem. Soc. Rev. 2014, 43, 5896–5912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhamid, H.N.; Dowaidar, M.; Hällbrink, M.; Langel, Ü. Gene Delivery Using Cell Penetrating Peptides-Zeolitic Imidazolate Frameworks. Microporous Mesoporous Mater. 2020, 300, 110173. [Google Scholar] [CrossRef]
- Qin, Y.-T.; Peng, H.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. pH-Responsive Polymer-Stabilized ZIF-8 Nanocomposites for Fluorescence and Magnetic Resonance Dual-Modal Imaging-Guided Chemo-/Photodynamic Combinational Cancer Therapy. ACS Appl. Mater. Interfaces 2019, 11, 34268–34281. [Google Scholar] [CrossRef]
- Vasconcelos, I.B.; da Silva, T.G.; Militão, G.C.G.; Soares, T.A.; Rodrigues, N.M.; Rodrigues, M.O.; da Costa, N.B.; Freire, R.O.; Junior, S.A. Cytotoxicity and Slow Release of the Anti-Cancer Drug Doxorubicin from ZIF-8. RSC Adv. 2012, 2, 9437–9442. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Zhang, W.; Xu, J.; Hou, J.; Feng, X.; Zhu, W. MOF Nanoparticles with Encapsulated Dihydroartemisinin as a Controlled Drug Delivery System for Enhanced Cancer Therapy and Mechanism Analysis. J. Mater. Chem. B Mater. Biol. Med. 2020, 8, 7382–7389. [Google Scholar] [CrossRef]
- Xie, R.; Yang, P.; Peng, S.; Cao, Y.; Yao, X.; Guo, S.; Yang, W. A Phosphorylcholine-Based Zwitterionic Copolymer Coated ZIF-8 Nanodrug with a Long Circulation Time and Charged Conversion for Enhanced Chemotherapy. J. Mater. Chem. B Mater. Biol. Med. 2020, 8, 6128–6138. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, L.; Yang, Y.; Qian, X.; Fu, T.; Li, X.; Yang, Z.; Yan, H.; Cui, C.; Tan, W. Metal-Organic Framework Nanocarriers for Drug Delivery in Biomedical Applications. Nanomicro Lett. 2020, 12, 103. [Google Scholar] [CrossRef]
- Feng, S.; Zhang, X.; Shi, D.; Wang, Z. Zeolitic Imidazolate Framework-8 (ZIF-8) for Drug Delivery: A Critical Review. Front. Chem. Sci. Eng. 2021, 15, 221–237. [Google Scholar] [CrossRef]
- Sun, C.-Y.; Qin, C.; Wang, X.-L.; Yang, G.-S.; Shao, K.-Z.; Lan, Y.-Q.; Su, Z.-M.; Huang, P.; Wang, C.-G.; Wang, E.-B. Zeolitic Imidazolate Framework-8 as Efficient pH-Sensitive Drug Delivery Vehicle. Dalton Trans. 2012, 41, 6906–6909. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Singh, A.; Garg, N.; Randhawa, J.K. Curcumin Encapsulated Zeolitic Imidazolate Frameworks as Stimuli Responsive Drug Delivery System and Their Interaction with Biomimetic Environment. Sci. Rep. 2017, 7, 12598. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ye, H.; Zhao, F.; Zeng, B. High-Quality Metal-Organic Framework ZIF-8 Membrane Supported on Electrodeposited ZnO/2-Methylimidazole Nanocomposite: Efficient Adsorbent for the Enrichment of Acidic Drugs. Sci. Rep. 2017, 7, 39778. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, C.; Das, A.; Chakraborty, A. Zeolitic Imidazole Framework (ZIF) Nanospheres for Easy Encapsulation and Controlled Release of an Anticancer Drug Doxorubicin under Different External Stimuli: A Way toward Smart Drug Delivery System. Mol. Pharm. 2015, 12, 3158–3166. [Google Scholar] [CrossRef]
- Chowdhury, M.A. The Applications of Metal-Organic-Frameworks in Controlled Release of Drugs. J. Macromol. Sci. Rev. Macromol. Chem. Phys. 2017, 7, 1–22. [Google Scholar] [CrossRef]
- Ettlinger, R.; Moreno, N.; Volkmer, D.; Kerl, K.; Bunzen, H. Zeolitic Imidazolate Framework-8 as pH-Sensitive Nanocarrier for “Arsenic Trioxide” Drug Delivery. Chemistry 2019, 25, 13189–13196. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Xu, W.; Meng, X.; Shi, X.; Shao, L.; Zeng, X.; Yang, Z.; Li, S.; Liu, Y.; Xia, X. A pH-Responsive Hydrophilic Controlled Release System Based on ZIF-8 for Self-Healing Anticorrosion Application. Chem. Eng. J. 2021, 415, 128985. [Google Scholar] [CrossRef]
- Luzuriaga, M.A.; Welch, R.P.; Dharmarwardana, M.; Benjamin, C.E.; Li, S.; Shahrivarkevishahi, A.; Popal, S.; Tuong, L.H.; Creswell, C.T.; Gassensmith, J.J. Enhanced Stability and Controlled Delivery of MOF-Encapsulated Vaccines and Their Immunogenic Response In Vivo. ACS Appl. Mater. Interfaces 2019, 11, 9740–9746. [Google Scholar] [CrossRef]
- Lin, Y.; Zhong, Y.; Chen, Y.; Li, L.; Chen, G.; Zhang, J.; Li, P.; Zhou, C.; Sun, Y.; Ma, Y.; et al. Ligand-Modified Erythrocyte Membrane-Cloaked Metal-Organic Framework Nanoparticles for Targeted Antitumor Therapy. Mol. Pharm. 2020, 17, 3328–3341. [Google Scholar] [CrossRef]
- Cheng, G.; Li, W.; Ha, L.; Han, X.; Hao, S.; Wan, Y.; Wang, Z.; Dong, F.; Zou, X.; Mao, Y.; et al. Self-Assembly of Extracellular Vesicle-like Metal-Organic Framework Nanoparticles for Protection and Intracellular Delivery of Biofunctional Proteins. J. Am. Chem. Soc. 2018, 140, 7282–7291. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater. 2016, 1, 1–12. [Google Scholar] [CrossRef]
- Liang, K.; Ricco, R.; Doherty, C.M.; Styles, M.J.; Bell, S.; Kirby, N.; Mudie, S.; Haylock, D.; Hill, A.J.; Doonan, C.J.; et al. Biomimetic Mineralization of Metal-Organic Frameworks as Protective Coatings for Biomacromolecules. Nat. Commun. 2015, 6, 7240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Sudlow, G.; Wang, Z.; Cao, S.; Jiang, Q.; Neiner, A.; Morrissey, J.J.; Kharasch, E.D.; Achilefu, S.; Singamaneni, S. Metal-Organic Framework Encapsulation Preserves the Bioactivity of Protein Therapeutics. Adv. Healthc. Mater. 2018, 7, e1800950. [Google Scholar] [CrossRef]
- Lyu, F.; Zhang, Y.; Zare, R.N.; Ge, J.; Liu, Z. One-Pot Synthesis of Protein-Embedded Metal-Organic Frameworks with Enhanced Biological Activities. Nano Lett. 2014, 14, 5761–5765. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.-S.; Lo, W.-S.; Hsu, Y.-S.; Wu, C.-C.; Wang, S.-C.; Shieh, F.-K.; Morabito, J.V.; Chou, L.-Y.; Wu, K.C.-W.; Tsung, C.-K. Shielding against Unfolding by Embedding Enzymes in Metal-Organic Frameworks via a de Novo Approach. J. Am. Chem. Soc. 2017, 139, 6530–6533. [Google Scholar] [CrossRef]
- Astria, E.; Thonhofer, M.; Ricco, R.; Liang, W.; Chemelli, A.; Tarzia, A.; Alt, K.; Hagemeyer, C.E.; Rattenberger, J.; Schroettner, H.; et al. Carbohydrates@MOFs. Mater. Horiz. 2019, 6, 969–977. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Xu, H.; Carraro, F.; Maddigan, N.K.; Li, Q.; Bell, S.G.; Huang, D.M.; Tarzia, A.; Solomon, M.B.; Amenitsch, H.; et al. Enhanced Activity of Enzymes Encapsulated in Hydrophilic Metal-Organic Frameworks. J. Am. Chem. Soc. 2019, 141, 2348–2355. [Google Scholar] [CrossRef]
- Ranji-Burachaloo, H.; Reyhani, A.; Gurr, P.A.; Dunstan, D.E.; Qiao, G.G. Combined Fenton and Starvation Therapies Using Hemoglobin and Glucose Oxidase. Nanoscale 2019, 11, 5705–5716. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, Y.; Liu, L.; Wan, W.; Guo, P.; Nyström, A.M.; Zou, X. One-Pot Synthesis of Metal-Organic Frameworks with Encapsulated Target Molecules and Their Applications for Controlled Drug Delivery. J. Am. Chem. Soc. 2016, 138, 962–968. [Google Scholar] [CrossRef]
- Chen, W.-H.; Luo, G.-F.; Vázquez-González, M.; Cazelles, R.; Sohn, Y.S.; Nechushtai, R.; Mandel, Y.; Willner, I. Glucose-Responsive Metal-Organic-Framework Nanoparticles Act as “Smart” Sense-and-Treat Carriers. ACS Nano 2018, 12, 7538–7545. [Google Scholar] [CrossRef]
- Li, S.; Dharmarwardana, M.; Welch, R.P.; Benjamin, C.E.; Shamir, A.M.; Nielsen, S.O.; Gassensmith, J.J. Investigation of Controlled Growth of Metal-Organic Frameworks on Anisotropic Virus Particles. ACS Appl. Mater. Interfaces 2018, 10, 18161–18169. [Google Scholar] [CrossRef] [PubMed]
- Riccò, R.; Liang, W.; Li, S.; Gassensmith, J.J.; Caruso, F.; Doonan, C.; Falcaro, P. Metal-Organic Frameworks for Cell and Virus Biology: A Perspective. ACS Nano 2018, 12, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhang, X.; Huang, L.; Zhang, Z.; Dong, S. GOx@ZIF-8(NiPd) Nanoflower: An Artificial Enzyme System for Tandem Catalysis. Angew. Chem. Weinheim Bergstr. Ger. 2017, 129, 16298–16301. [Google Scholar] [CrossRef]
- Liang, W.; Ricco, R.; Maddigan, N.K.; Dickinson, R.P.; Xu, H.; Li, Q.; Sumby, C.J.; Bell, S.G.; Falcaro, P.; Doonan, C.J. Control of Structure Topology and Spatial Distribution of Biomacromolecules in Protein@ZIF-8 Biocomposites. Chem. Mater. 2018, 30, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Zhang, L.; An, J.; Wang, T.; Li, L.; Si, X.; He, L.; Wu, X.; Wang, C.; Su, Z. Polyacrylic Acid@zeolitic Imidazolate Framework-8 Nanoparticles with Ultrahigh Drug Loading Capability for pH-Sensitive Drug Release. Chem. Commun. 2014, 50, 1000–1002. [Google Scholar] [CrossRef]
- Bian, R.; Wang, T.; Zhang, L.; Li, L.; Wang, C. A Combination of Tri-Modal Cancer Imaging and in Vivo Drug Delivery by Metal-Organic Framework Based Composite Nanoparticles. Biomater. Sci. 2015, 3, 1270–1278. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Gong, K.; Chen, J. Nanoscale Zeolitic Imidazolate Framework-8 as Efficient Vehicles for Enhanced Delivery of CpG Oligodeoxynucleotides. ACS Appl. Mater. Interfaces 2017, 9, 31519–31525. [Google Scholar] [CrossRef]
- Silva, J.S.F.; Silva, J.Y.R.; de Sá, G.F.; Araújo, S.S.; Filho, M.A.G.; Ronconi, C.M.; Santos, T.C.; Júnior, S.A. Multifunctional System Polyaniline-Decorated ZIF-8 Nanoparticles as a New Chemo-Photothermal Platform for Cancer Therapy. ACS Omega 2018, 3, 12147–12157. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; You, Y.; Zeng, F.; Wang, Y.; Liang, C.; Feng, H.; Ma, X. Disassembly of Hydrophobic Photosensitizer by Biodegradable Zeolitic Imidazolate Framework-8 for Photodynamic Cancer Therapy. ACS Appl. Mater. Interfaces 2018, 10, 15517–15523. [Google Scholar] [CrossRef]
- Anai, C.; Kawaguchi, M.; Eto, K. Effects of Culture Media on the Susceptibility of Cells to Apoptotic Cell Death. Vitr. Cell. Dev. Biol. Anim. 2014, 50, 683–687. [Google Scholar] [CrossRef]
- Patel, V.; Amin, K.; Allen, D.; Ukishima, L.; Wahab, A.; Grodi, C.; Behrsing, H. Comparison of Long-Term Human Precision-Cut Lung Slice Culture Methodology and Response to Challenge: An Argument for Standardisation. Altern. Lab. Anim. 2021, 49, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Velásquez-Hernández, M.D.J.; Ricco, R.; Carraro, F.; Limpoco, F.T.; Linares-Moreau, M.; Leitner, E.; Wiltsche, H.; Rattenberger, J.; Schröttner, H.; Frühwirt, P.; et al. Degradation of ZIF-8 in Phosphate Buffered Saline Media. CrystEngComm 2019, 21, 4538–4544. [Google Scholar] [CrossRef] [Green Version]
- Poryvaev, A.S.; Yazikova, A.A.; Polyukhov, D.M.; Chinak, O.A.; Richter, V.A.; Krumkacheva, O.A.; Fedin, M.V. Guest Leakage from ZIF-8 Particles under Drug Delivery Conditions: Quantitative Characterization and Guest-Induced Framework Stabilization. J. Phys. Chem. C 2021, 125, 15606–15613. [Google Scholar] [CrossRef]
- Luzuriaga, M.A.; Benjamin, C.E.; Gaertner, M.W.; Lee, H.; Herbert, F.C.; Mallick, S.; Gassensmith, J.J. ZIF-8 Degrades in Cell Media, Serum, and Some-But Not All-Common Laboratory Buffers. Supramol. Chem. 2019, 31, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Ashok, D.; Sen, T.; Enge, T.G.; Verma, N.K.; Tricoli, A.; Lowe, A.; Nisbet, R.D.; Tsuzuki, T. Stability of ZIF-8 Nanopowders in Bacterial Culture Media and Its Implication for Antibacterial Properties. Chem. Eng. J. 2021, 413, 127511. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, S.; Guan, X.; Xie, Z. One-Step Synthesis of Nanoscale Zeolitic Imidazolate Frameworks with High Curcumin Loading for Treatment of Cervical Cancer. ACS Appl. Mater. Interfaces 2015, 7, 22181–22187. [Google Scholar] [CrossRef]
- Chen, P.; He, M.; Chen, B.; Hu, B. Size- and Dose-Dependent Cytotoxicity of ZIF-8 Based on Single Cell Analysis. Ecotoxicol. Environ. Saf. 2020, 205, 111110. [Google Scholar] [CrossRef]
- Ruyra, À.; Yazdi, A.; Espín, J.; Carné-Sánchez, A.; Roher, N.; Lorenzo, J.; Imaz, I.; Maspoch, D. Synthesis, Culture Medium Stability, and in Vitro and in Vivo Zebrafish Embryo Toxicity of Metal-Organic Framework Nanoparticles. Chemistry 2015, 21, 2508–2518. [Google Scholar] [CrossRef]
- Jiang, Z.; Wang, Y.; Sun, L.; Yuan, B.; Tian, Y.; Xiang, L.; Li, Y.; Li, Y.; Li, J.; Wu, A. Dual ATP and pH Responsive ZIF-90 Nanosystem with Favorable Biocompatibility and Facile Post-Modification Improves Therapeutic Outcomes of Triple Negative Breast Cancer in Vivo. Biomaterials 2019, 197, 41–50. [Google Scholar] [CrossRef]
- Dobrynin, S.A.; Glazachev, Y.I.; Gatilov, Y.V.; Chernyak, E.I.; Salnikov, G.E.; Kirilyuk, I.A. Synthesis of 3,4-Bis(hydroxymethyl)-2,2,5,5-Tetraethylpyrrolidin-1-Oxyl via 1,3-Dipolar Cycloaddition of Azomethine Ylide to Activated Alkene. J. Org. Chem. 2018, 83, 5392–5397. [Google Scholar] [CrossRef]
- Polyukhov, D.M.; Poryvaev, A.S.; Gromilov, S.A.; Fedin, M.V. Precise Measurement and Controlled Tuning of Effective Window Sizes in ZIF-8 Framework for Efficient Separation of Xylenes. Nano Lett. 2019, 19, 6506–6510. [Google Scholar] [CrossRef] [PubMed]
- Simon-Yarza, T.; Mielcarek, A.; Couvreur, P.; Serre, C. Nanoparticles of Metal-Organic Frameworks: On the Road to In Vivo Efficacy in Biomedicine. Adv. Mater. 2018, 30, e1707365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, D.; Yao, Y.; Zhang, B.; Lin, Y.S. Stability of ZIF-8 Membranes and Crystalline Powders in Water at Room Temperature. J. Memb. Sci. 2015, 485, 103–111. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, M.; Lin, Y.S. Stability of ZIF-8 in Water under Ambient Conditions. Microporous Mesoporous Mater. 2019, 279, 201–210. [Google Scholar] [CrossRef]
- Hallman, P.S.; Perrin, D.D.; Watt, A.E. The Computed Distribution of copper(II) and zinc(II) Ions among Seventeen Amino Acids Present in Human Blood Plasma. Biochem. J. 1971, 121, 549–555. [Google Scholar] [CrossRef]
- Abendrot, M.; Chęcińska, L.; Kusz, J.; Lisowska, K.; Zawadzka, K.; Felczak, A.; Kalinowska-Lis, U. Zinc(II) Complexes with Amino Acids for Potential Use in Dermatology: Synthesis, Crystal Structures, and Antibacterial Activity. Molecules 2020, 25, 951. [Google Scholar] [CrossRef] [Green Version]
- Pace, N.J.; Weerapana, E. Zinc-Binding Cysteines: Diverse Functions and Structural Motifs. Biomolecules 2014, 4, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Bell, P.; Sheldrick, W.S. Preparation and Structure of Zinc Complexes of Cysteine Derivatives. Z. Für. Nat. B 1984, 39, 1732–1737. [Google Scholar] [CrossRef] [Green Version]
- Gaizer, F.; Silber, H.B. Stability Constants of Zinc Chloride Complexes in DMSO-Water Mixtures. J. Inorg. Nucl. Chem. 1980, 42, 1317–1320. [Google Scholar] [CrossRef]
- Stoll, S.; Schweiger, A. EasySpin, a Comprehensive Software Package for Spectral Simulation and Analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spitsyna, A.S.; Poryvaev, A.S.; Sannikova, N.E.; Yazikova, A.A.; Kirilyuk, I.A.; Dobrynin, S.A.; Chinak, O.A.; Fedin, M.V.; Krumkacheva, O.A. Stability of ZIF-8 Nanoparticles in Most Common Cell Culture Media. Molecules 2022, 27, 3240. https://doi.org/10.3390/molecules27103240
Spitsyna AS, Poryvaev AS, Sannikova NE, Yazikova AA, Kirilyuk IA, Dobrynin SA, Chinak OA, Fedin MV, Krumkacheva OA. Stability of ZIF-8 Nanoparticles in Most Common Cell Culture Media. Molecules. 2022; 27(10):3240. https://doi.org/10.3390/molecules27103240
Chicago/Turabian StyleSpitsyna, Anna S., Artem S. Poryvaev, Natalya E. Sannikova, Anastasiya A. Yazikova, Igor A. Kirilyuk, Sergey A. Dobrynin, Olga A. Chinak, Matvey V. Fedin, and Olesya A. Krumkacheva. 2022. "Stability of ZIF-8 Nanoparticles in Most Common Cell Culture Media" Molecules 27, no. 10: 3240. https://doi.org/10.3390/molecules27103240