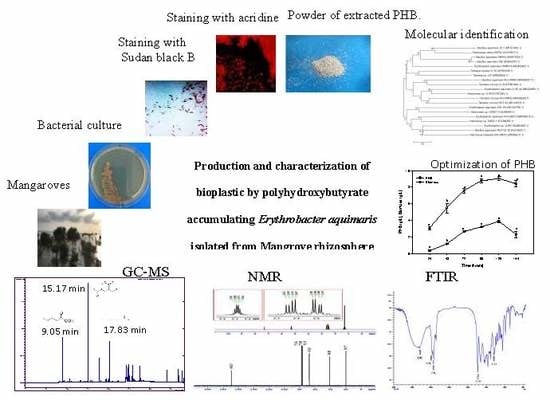
Production and Characterization of Bioplastic by Polyhydroxybutyrate Accumulating Erythrobacter aquimaris Isolated from Mangrove Rhizosphere
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Marine Bacteria
2.2. Qualitative Screening and Selection of Poly-β-hydroxybutyrate-Producing Marine Bacteria
2.3. Bacterial Identification Using 16S rRNA Gene and Phylogenetic Analysis
2.4. Quantitative Screening of PHB-Producing Marine Bacteria
2.5. Optimization of PHB Production by E. Aquimaris
2.5.1. Effect of Fermentation Period on PHB Production
2.5.2. Effect of Initial pH on PHB Production
2.5.3. Effect of Incubation Temperature on PHB Production
2.5.4. Effect of NaCl Concentration on PHB Production
2.5.5. Effect of Carbon Sources on PHB Production
2.5.6. Effect of Nitrogen Sources on PHB Production
2.5.7. Effect of C:N Ratio on the PHB Production
2.6. Chemical Characterization of Extracted PHB
2.6.1. Fourier-Transform Infrared Spectroscopy Analysis
2.6.2. Nuclear Magnetic Resonance Spectroscopy Analysis
2.6.3. Gas Chromatography-Mass Spectrometry Analysis
3. Materials and Methods
3.1. Sample Collection
3.2. Isolation of PHB-Producing Marine Bacteria
3.3. Screening of PHB-Producing Bacteria
3.3.1. Screening by Sudan Black-B Stain
3.3.2. Screening by Fluorescence Staining Method
3.3.3. Quantitative Screening of PHB Production
3.4. Extraction and Quantitative Estimation of PHB
3.5. Molecular Identification of the Promising PHB-Producing Isolates
3.5.1. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification of 16S rRNA Gene
3.5.2. Sequencing and GenBank Accession Numbers
3.6. Optimization of PHB Production
3.7. Characterization of Extracted PHB
3.7.1. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
3.7.2. Nuclear Magnetic Resonance (NMR) Analysis
3.7.3. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, P.; Sharma, V. Integrated Plastic Waste Management: Environmental and Improved Health Approaches. Procedia Environ. Sci. 2016, 35, 692–700. [Google Scholar] [CrossRef]
- Mittal, N.; Jansson, R.; Widhe, M.; Benselfelt, T.; Håkansson, K.M.O.; Lundell, F.; Hedhammar, M.; Söderberg, L.D. Ultrastrong and Bioactive Nanostructured Bio-Based Composites. ACS Nano 2017, 11, 5148–5159. [Google Scholar] [CrossRef]
- Mittal, N.; Ansari, F.; Gowda, V.K.; Brouzet, C.; Chen, P.; Larsson, P.T.; Roth, S.V.; Lundell, F.; Wågberg, L.; Kotov, N.A.; et al. Multiscale Control of Nanocellulose Assembly: Transferring Remarkable Nanoscale Fibril Mechanics to Macroscale Fibers. ACS Nano 2018, 12, 6378–6388. [Google Scholar] [CrossRef]
- Mittal, N.; Benselfelt, T.; Ansari, F.; Gordeyeva, K.; Roth, S.V.; Wågberg, L.; Söderberg, L.D.; Söderberg, D. Ion-Specific Assembly of Strong, Tough, and Stiff Biofibers. Angew. Chem. Int. Ed. 2019, 58, 18562–18569. [Google Scholar] [CrossRef] [Green Version]
- Mishra, G.; Mittal, N.; Sharma, A. Multifunctional mesoporous carbon capsules and their robust coatings for encapsulation of actives: Antimicrobial and anti-bioadhesion functions. Appl. Mater. Interfaces. 2017, 9, 23. [Google Scholar] [CrossRef]
- Alzubaidy, H.; Essack, M.; Malas, T.B.; Bokhari, A.; Motwalli, O.; Kamanu, F.K.; Jamhor, S.A.; Mokhtar, N.A.; Antunes, A.; Simões, M.F.; et al. Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene 2016, 576, 626–636. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.-W.; Song, H.-S.; Moon, Y.-M.; Hong, Y.-G.; Bhatia, S.K.; Jung, H.-R.; Choi, T.-R.; Yang, S.-Y.; Park, H.-Y.; Choi, Y.-K.; et al. Polyhydroxybutyrate production in halophilic marine bacteria Vibrio proteolyticus isolated from the Korean peninsula. Bioprocess Biosyst. Eng. 2019, 42, 603–610. [Google Scholar] [CrossRef]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioeng 2017, 4, 55. [Google Scholar] [CrossRef] [Green Version]
- Chien, C.-C.; Chen, C.-C.; Choi, M.-H.; Kung, S.-S.; Wei, Y.-H. Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J. Biotechnol. 2007, 132, 259–263. [Google Scholar] [CrossRef]
- Tan, G.-Y.A.; Chen, C.-L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.-Y. Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef] [Green Version]
- Możejko-Ciesielska, J.; Kiewisz, R. Bacterial polyhydroxyalkanoates: Still fabulous? Microbiol. Res. 2016, 192, 271–282. [Google Scholar] [CrossRef]
- Castro-Mayorga, J.; Freitas, F.; Reis, M.; Prieto, M.; Lagaron, J.; Prieto, A. Biosynthesis of silver nanoparticles and polyhydroxybutyrate nanocomposites of interest in antimicrobial applications. Int. J. Boil. Macromol. 2018, 108, 426–435. [Google Scholar] [CrossRef]
- Kalia, V.C.; Ray, S.; Patel, S.K.; Singh, M.; Singh, G.P. Biotechnological Applications of Polyhydroxyalkanoates: Applications of Polyhydroxyalkanoates and Their Metabolites as Drug Carriers; Springer: Singapore, 2019. [Google Scholar]
- Juengert, J.; Bresan, S.; Jendrossek, D. Determination of Polyhydroxybutyrate (PHB) Content in Ralstonia eutropha Using Gas Chromatography and Nile Red Staining. Bio-Protocol. 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.-L.; Wu, Y.-H.; Jin, X.-B.; Wang, C.-S.; Xu, X.-W. Alteromonas lipolytica sp. nov., a poly-beta-hydroxybutyrate-producing bacterium isolated from surface seawater. Int. J. Syst. Evol. Microbiol. 2017, 67, 237–242. [Google Scholar] [CrossRef]
- Tufail, S.; Munir, S.; Jamil, N. Variation analysis of bacterial polyhydroxyalkanoates production using saturated and unsaturated hydrocarbons. Braz. J. Microbiol. 2017, 48, 629–636. [Google Scholar] [CrossRef]
- Gabr, G.A. Isolation and identification of bacterial strains able to biopolymer polyhydroxybutyrate (Phb) production from soil of Al-Kharj probes, Saudi Arabia. J. Pharm. Res. Inter. 2019, 21. [Google Scholar] [CrossRef]
- Sasidharan, R.S.; Bhat, S.G.; Chandrasekaran, M. Biocompatible polyhydroxybutyrate (PHB) production by marine Vibrio azureus BTKB33 under submerged fermentation. Ann. Microbiol. 2015, 65, 455–465. [Google Scholar] [CrossRef]
- Nielsen, J.; Archer, J.; Essack, M.; Bajic, V.B.; Gojobori, T.; Mijakovic, I. Building a bio-based industry in the Middle East through harnessing the potential of the Red Sea biodiversity. Appl. Microbiol. Biotechnol. 2017, 101, 4837–4851. [Google Scholar] [CrossRef] [Green Version]
- Thompson, L.R.; Williams, G.J.; Haroon, M.F.; Shibl, A.; Larsen, P.; Shorenstein, J.; Knight, R.; Stingl, U. Metagenomic covariation along densely sampled environmental gradients in the Red Sea. ISME J. 2017, 11, 138–151. [Google Scholar] [CrossRef] [Green Version]
- Sholkamy, E.N.; Munusamy, M.A.; Alarfaj, A.A.; Arshad, M. Extraction and characterization of polyhydroxybutyrates (PHB) from Bacillus thuringiensis KSADL127 isolated from Mangrove environments of Saudi Arabia. Braz. Arch. Boil. Technol. 2015, 58, 781–788. [Google Scholar]
- Moorkoth, D.; Nampoothiri, K.M. Production and characterization of poly (3-hydroxy butyrate-co-3 hydroxyvalerate) (PHBV) by a novel halotolerant mangrove isolate. Bioresour. Technol. 2016, 201, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, R.; Hill, D.; Kenward, M.; Williams, C.; Radecka, I. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 2007, 102, 1437–1449. [Google Scholar] [CrossRef]
- Devi, A.B.; Nachiyar, C.V.; Kaviyarasi, T.; Samrot, A.V. Characterization of polyhydroxybutyrate synthesized by Bacillus cereus. Int. J. Pharm. Pharm. Sci. 2015, 7, 140–144. [Google Scholar]
- Bhagowati, P.; Pradhan, S.; Dash, H.R.; Das, S. Production, optimization and characterization of polyhydroxybutyrate, a biodegradable plastic by Bacillus spp. Biosci. Biorech. Bioch. 2015, 79, 1454–1463. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Kumar, A.; Muqtadir, A. Distribution of mangroves along the Red Sea coast of the Arabian Peninsula: Part 2. The southern coast of western Saudi Arabia. J. Earth Sci. India. 2010, 3, 154–162. [Google Scholar]
- Mahitha, G.; Madhuri, R. Purification and characterization of polyhydroxybutyrate produced from marine bacteria. Int. J. Sci. Eng. Res. 2015, 6, 71–75. [Google Scholar]
- Kalaivani, R.; Sukumaran, V. Isolation and identification of new strains to enhance the production of biopolymers from marine sample in Karankura, Tamil Nadu. Euro. J. Exp. Bio. 2013, 3, 56–64. [Google Scholar]
- Burdon, K.L. Fatty material in bacteria and fungi revealed by staining dried, fixed slide preparations. J. Bacteriol. 1946, 52, 665–678. [Google Scholar]
- Lorch, E.; Rentsch, G. A simple method for staining and counting isolated adipose tissue fat cells. Diabetol. 1969, 5, 356–357. [Google Scholar] [CrossRef] [Green Version]
- Qian, P.-Y.; Wang, Y.; Lee, O.O.; Lau, S.C.K.; Yang, J.; Lafi, F.F.; Al-Suwailem, A.; Wong, T.Y.H. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 2011, 5, 568. [Google Scholar] [CrossRef]
- Quillaguamán, J.; Guzmán, H.; Van-Thuoc, D.; Hatti-Kaul, R. Synthesis and production of polyhydroxyalkanoates by halophiles: Current potential and future prospects. Appl. Microbiol. Biotechnol. 2010, 85, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Arun, A.; Arthi, R.; Shanmugabalaji, V.; Eyini, M. Microbial production of poly-β-hydroxybutyrate by marine microbes isolated from various marine environments. Bioresour. Technol. 2009, 100, 2320–2323. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Production of polyhydroxyalkanoates (PHAs) by Bacillus subtilis and Escherichia coli grown on cane molasses fortified with ethanol. Braz. Arch. Boil. Technol. 2014, 57, 145–154. [Google Scholar] [CrossRef]
- Belal, E.B. Production of poly-β-Hydroxybutyric acid (PHB) by Rhizobium elti and Pseudomonas stutzeri. Curr. Res. J. Biol. Sci. 2013, 5, 273–284. [Google Scholar] [CrossRef]
- Lathwal, P.; Nehra, K.; Singh, M.; Jamdagani, P.; Rana, J.S. Optimization of culture parameters for maximum polyhydroxybutyrate production by selected bacterial strains isolated from rhizospheric soils. Pol. J. Microbiol. 2015, 64, 227–239. [Google Scholar] [CrossRef]
- Sathiyanarayanan, G.; Saibaba, G.; Kiran, G.S.; Selvin, J. A statistical approach for optimization of polyhydroxybutyrate production by marine Bacillus subtilis MSBN17. Int. J. Boil. Macromol. 2013, 59, 170–177. [Google Scholar] [CrossRef]
- Dhangdhariya, J.H.; Dubey, S.; Trivedi, H.B.; Pancha, I.; Bhatt, J.K.; Dave, B.P.; Mishra, S. Polyhydroxyalkanoate from marine Bacillus megaterium using CSMCRI’s dry sea mix as a novel growth medium. Int. J. Biol. Macromol. 2015, 76, 254–261. [Google Scholar] [CrossRef]
- Omar, S.; Rayes, A.; Eqaab, A.; Vob, I.; Steinbüchel, A. Optimization of cell growth and poly(3-hydroxybutyrate) accumulation on date syrup by a Bacillus megaterium strain. Biotech. Lett. 2011, 23, 1119–1123. [Google Scholar] [CrossRef]
- Mohandas, S.; Balan, L.; Lekshmi, N.; Cubelio, S.; Philip, R.; Bright Singh, I. Production and characterization of polyhydroxybutyrate from Vibrio harveyi MCCB 284 utilizing glycerol as carbon source. J. Appl. Microbiol. 2017, 122, 698–707. [Google Scholar] [CrossRef]
- Getachew, A.; Woldesenbet, F. Production of biodegradable plastic by polyhydroxybutyrate (PHB) accumulating bacteria using low cost agricultural waste material. BMC Res. Notes 2016, 9, 509. [Google Scholar] [CrossRef] [Green Version]
- Maheshwari, D.K.; Saraf, M. Halophiles: Biodiversity and Sustainable Exploitation; Springer International Publishing: Basel, Switzerland, 2015. [Google Scholar] [CrossRef]
- Santhanam, A.; Sasidharan, S. Microbial production of polyhydroxy alkanotes (PHA) from Alcaligens spp. and Pseudomonas oleovorans using different carbon sources. Afr. J. Biotechnol. 2010, 9, 3144–3150. [Google Scholar]
- Khiyami, M.A.; Al-Fadual, S.M.; Bahklia, A.H. Polyhydroxyalkanoates production via Bacillus plastic composite support (PCS) biofilm and date palm syrup. J. Med. Plant. Res. 2011, 5, 3312–3320. [Google Scholar]
- Cardozo, J.R.G.; Martínez, A.L.M.; Pérez, M.Y.; Londoño, G.A.C. Production and characterization of polyhydroxyalkanoates and native microorganisms synthesized from fatty waste. Int. J. Polym. Sci. 2016, 2016, 6541718. [Google Scholar] [CrossRef] [Green Version]
- Hungund, B.; Vs, S.; Patwardhan, P.; Saleh, A.M. Production of Polyhydroxyalkanoate from Paenibacillus durus BV-1 Isolated from Oil Mill Soil. J. Microb. Biochem. Technol. 2013, 5, 013–017. [Google Scholar] [CrossRef] [Green Version]
- Patel, N.; Patel, P.; Desai, R. Detection & characterization of PHB (polyhydroxybutyrate) Producers halophilic bacteria isolated from marine water sample of Valsad District. Int. J. Pharma Bio Sci. 2017, 8, 1100. [Google Scholar]
- Gumel, A.M.; Annuar, M.S.M.; Heidelberg, T. Biosynthesis and characterization of polyhydroxyalkanoates copolymers produced by Pseudomonas putida Bet001 isolated from Palm Oil mill effluent. PLoS ONE 2012, 7, e45214. [Google Scholar] [CrossRef] [Green Version]
- Likitha, R.V.; Namratha, V.; Sarada, B. Production of PHB from Cupriavidus necator using Calophyllum inophyllum seed oil as carbon source. Int. J. Life. Sci. Res. 2018, 6, 208–2011. [Google Scholar]
- Biradar, G.G.; Shivasharana, C.; Kaliwal, B.B. Characterization of polyhydroxybutyrate (PHB) produced by novel bacterium Lysinibacillus sphaericus bbkgbs6 isolated from soil. J. Polym. Environ. 2018, 26, 1685–1701. [Google Scholar] [CrossRef]
- Yang, C.; He, N.; Ling, X.; Ye, M.; Zhang, C.; Shao, W.; Yao, C.; Wang, Z.; Li, Q. The isolation and characterization of polysaccharides from longan pulp. Sep. Purif. Technol. 2008, 63, 226–230. [Google Scholar] [CrossRef]
- Shrivastav, A.; Mishra, S.K.; Shethia, B.; Pancha, I.; Jain, D.; Mishra, S. Isolation of promising bacterial strains from soil and marine environment for polyhydroxyalkanoates (PHAs) production utilizing Jatropha biodiesel byproduct. Int. J. Boil. Macromol. 2010, 47, 283–287. [Google Scholar] [CrossRef]
- Krishnan, S.; Chinnadurai, G.S.; Perumal, P. Polyhydroxybutyrate by Streptomyces sp.: Production and characterization. Int. J. Boil. Macromol. 2017, 104, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Neilan, B.A. Identification and phylogenetic analysis of toxigenic cyanobacteria by multiplex randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 1995, 61, 2286–2291. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |














| Isolates ID | Isolates Name | Similarity of 16S rRNA Gene (%) | Genbank Accession Number |
|---|---|---|---|
| KKU-MR35 | Tamlana crocina | 99.69% | MH266206.1 |
| KKU-MR42 | Bacillus aquimaris | 99.43% | MH266207.1 |
| KKU-MD13 | Erythrobacter aquimaris | 99.39% | MH266210.1 |
| KKU-MD12 | Halomonas halophila | 98.98% | MH266211.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa, Y.S.; Alrumman, S.A.; Otaif, K.A.; Alamri, S.A.; Mostafa, M.S.; Sahlabji, T. Production and Characterization of Bioplastic by Polyhydroxybutyrate Accumulating Erythrobacter aquimaris Isolated from Mangrove Rhizosphere. Molecules 2020, 25, 179. https://doi.org/10.3390/molecules25010179
Mostafa YS, Alrumman SA, Otaif KA, Alamri SA, Mostafa MS, Sahlabji T. Production and Characterization of Bioplastic by Polyhydroxybutyrate Accumulating Erythrobacter aquimaris Isolated from Mangrove Rhizosphere. Molecules. 2020; 25(1):179. https://doi.org/10.3390/molecules25010179
Chicago/Turabian StyleMostafa, Yasser S., Sulaiman A. Alrumman, Kholod A. Otaif, Saad A. Alamri, Mohamed S. Mostafa, and Taher Sahlabji. 2020. "Production and Characterization of Bioplastic by Polyhydroxybutyrate Accumulating Erythrobacter aquimaris Isolated from Mangrove Rhizosphere" Molecules 25, no. 1: 179. https://doi.org/10.3390/molecules25010179