Recent Advances in Tumor Suppressor (Closed)
A topical collection in Cancers (ISSN 2072-6694). This collection belongs to the section "Molecular Cancer Biology".
Viewed by 18316Editor
Interests: oncogene; tumor suppressor; tumor metabolism; RB; Ras; RECK
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
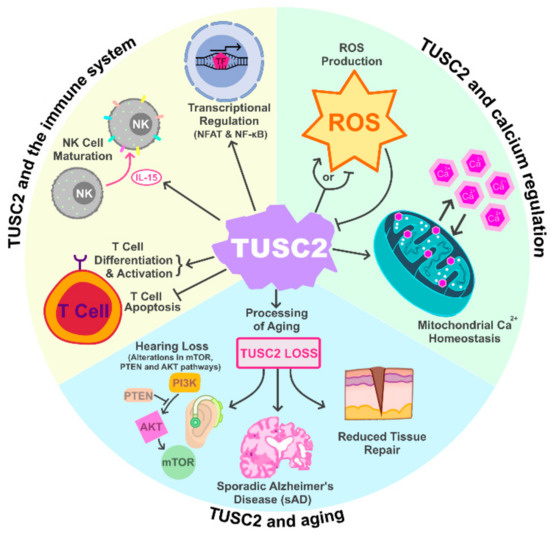
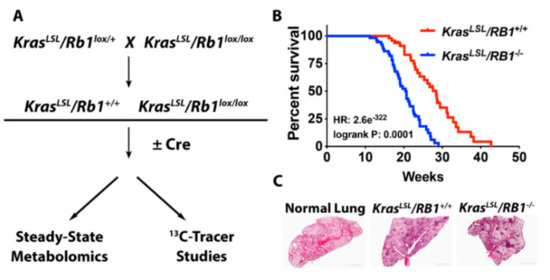
The era of investigations on tumor suppressor genes was initiated by the discovery of retinoblastoma tumor suppressor 1 gene (RB1) and Trp53 tumor suppressor gene that took place more than three decades ago. RB1 primarily regulates G1/S transition during cell cycle progression by modulating the activity of E2F transcription factors. RB mutation was initially discovered by virtue of its role in tumor initiation.
However, it is becoming clear that, in the majority of cancers, somatic RB1 inactivation occurs rather during tumor progression. The consequence of RB1 inactivation in this context contains epithelial mesenchymal transition (EMT), invasion, metastasis, undifferentiated status, tumor microenvironment, therapy resistance, etc.
As is in case of RB1, recent studies uncovered numerous novel functions in tumor suppressors that were unexpected in early studies. The aim of this Special Issue of Cancers is to highlight studies focusing on previously unexpected functions of various tumor suppressors including RB1. We welcome submissions that will contribute to deepen our understanding of cancers from the view of the complicated functions of tumor suppressors.
Prof. Dr. Chiaki TakahashiGuest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- retinoblastoma
- RB
- E2F
- cell cycle
- cancer progression