Identification of Proteins Specifically Assembled on a Stem-Loop Composed of a CAG Triplet Repeat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Plasmid DNA Substrates
2.3. TFO-Conjugated Plasmid on Magnetic Bead
2.4. Xenopus Egg Extracts
2.5. Plasmid DNA Pull-Down and Protein Analysis
3. Results
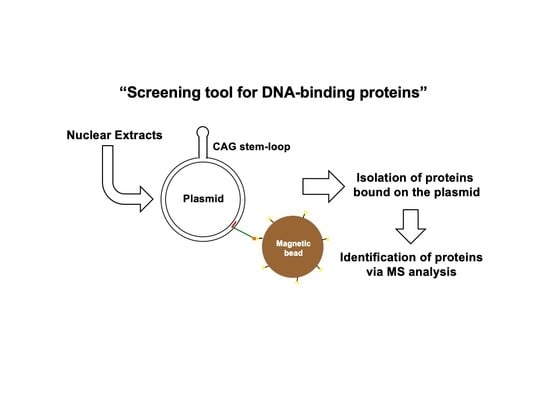
3.1. Assay Design to Capture and Identify Proteins Specifically Assembled on Structural DNA
3.2. Protein Capture in the Context of the Plasmid DNA Pull-Down Approach
3.3. Identification of Proteins Specifically Recruited on the Stem Loop
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.J.; Shaw, P.G.; Kim, M.-S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 Nucleotide Repeat Structures Initiate Molecular Cascades of Disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Yum, K.; Wang, E.T.; Kalsotra, A. Myotonic Dystrophy: Disease Repeat Range, Penetrance, Age of Onset, and Relationship between Repeat Size and Phenotypes. Curr. Opin. Genet. Dev. 2017, 44, 30–37. [Google Scholar] [CrossRef]
- Iyer, R.R.; Pluciennik, A.; Napierala, M.; Wells, R.D. DNA Triplet Repeat Expansion and Mismatch Repair. Annu. Rev. Biochem. 2015, 84, 199–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-M.; Correia, K.; Loupe, J.; Kim, K.-H.; Barker, D.; Hong, E.P.; Chao, M.J.; Long, J.D.; Lucente, D.; Vonsattel, J.P.G.; et al. CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell 2019, 178, 887–900.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovtun, I.V.; Liu, Y.; Bjoras, M.; Klungland, A.; Wilson, S.H.; McMurray, C.T. OGG1 Initiates Age-Dependent CAG Trinucleotide Expansion in Somatic Cells. Nature 2007, 447, 447–452. [Google Scholar] [CrossRef] [Green Version]
- Benn, C.L.; Gibson, K.R.; Reynolds, D.S. Drugging DNA Damage Repair Pathways for Trinucleotide Repeat Expansion Diseases. JHD 2021, 10, 203–220. [Google Scholar] [CrossRef]
- Wheeler, V.C.; Dion, V. Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models. JHD 2021, 10, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A. DNA Mismatch Repair and Its Role in Huntington’s Disease. JHD 2021, 10, 75–94. [Google Scholar] [CrossRef]
- Deshmukh, A.L.; Porro, A.; Mohiuddin, M.; Lanni, S.; Panigrahi, G.B.; Caron, M.-C.; Masson, J.-Y.; Sartori, A.A.; Pearson, C.E. FAN1, a DNA Repair Nuclease, as a Modifier of Repeat Expansion Disorders. JHD 2021, 10, 95–122. [Google Scholar] [CrossRef]
- Isogawa, A.; Fuchs, R.P.; Fujii, S. Versatile and Efficient Chromatin Pull-down Methodology Based on DNA Triple Helix Formation. Sci. Rep 2018, 8, 5925. [Google Scholar] [CrossRef] [Green Version]
- Isogawa, A.; Fuchs, R.P.; Fujii, S. Chromatin pull-down methodology based on DNA triple helix formation. In DNA Electrophoresis; Methods in Molecular Biology; Hanada, K., Ed.; Springer: New York, NY, USA, 2020; Volume 2119, pp. 183–199. [Google Scholar] [CrossRef]
- Cupello, S.; Richardson, C.; Yan, S. Cell-Free Xenopus Egg Extracts for Studying DNA Damage Response Pathways. Int. J. Dev. Biol. 2016, 60, 229–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, P.J.; Gambus, A.; Blow, J.J. Preparation and Use of Xenopus Egg Extracts to Study DNA Replication and Chromatin Associated Proteins. Methods 2012, 57, 203–213. [Google Scholar] [CrossRef]
- Wu, R.A.; Semlow, D.R.; Kamimae-Lanning, A.N.; Kochenova, O.V.; Chistol, G.; Hodskinson, M.R.; Amunugama, R.; Sparks, J.L.; Wang, M.; Deng, L.; et al. TRAIP Is a Master Regulator of DNA Interstrand Crosslink Repair. Nature 2019, 567, 267–272. [Google Scholar] [CrossRef]
- Fujii, S.; Sobol, R.W.; Fuchs, R.P. Double-Strand Breaks: When DNA Repair Events Accidentally Meet. DNA Repair 2022, 112, 103303. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, R.P.; Isogawa, A.; Paulo, J.A.; Onizuka, K.; Takahashi, T.; Amunugama, R.; Duxin, J.P.; Fujii, S. Crosstalk between Repair Pathways Elicits Double-Strand Breaks in Alkylated DNA and Implications for the Action of Temozolomide. eLife 2021, 10, e69544. [Google Scholar] [CrossRef] [PubMed]
- Richard, G.-F.; Kerrest, A.; Dujon, B. Comparative Genomics and Molecular Dynamics of DNA Repeats in Eukaryotes. Microbiol. Mol. Biol. Rev. 2008, 72, 686–727. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.; Sun, L.; Newport, J. Regulated Chromosomal DNA Replication in the Absence of a Nucleus. Mol. Cell 1998, 1, 519–529. [Google Scholar] [CrossRef]
- Lebofsky, R.; Takahashi, T.; Walter, J.C. DNA replication in nucleus-free Xenopus egg extracts. In DNA Replication; Methods in Molecular Biology; Vengrova, S., Dalgaard, J.Z., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 521, pp. 229–252. [Google Scholar] [CrossRef]
- Nakamori, M.; Panigrahi, G.B.; Lanni, S.; Gall-Duncan, T.; Hayakawa, H.; Tanaka, H.; Luo, J.; Otabe, T.; Li, J.; Sakata, A.; et al. A Slipped-CAG DNA-Binding Small Molecule Induces Trinucleotide-Repeat Contractions In Vivo. Nat. Genet. 2020, 52, 146–159. [Google Scholar] [CrossRef]
- Jiricny, J. The Multifaceted Mismatch-Repair System. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef]
- Mosbach, V.; Poggi, L.; Viterbo, D.; Charpentier, M.; Richard, G.-F. TALEN-Induced Double-Strand Break Repair of CTG Trinucleotide Repeats. Cell Rep. 2018, 22, 2146–2159. [Google Scholar] [CrossRef] [Green Version]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA Double-Strand Break Repair-Pathway Choice in Somatic Mammalian Cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Semlow, D.R.; Walter, J.C. Mechanisms of Vertebrate DNA Interstrand Cross-Link Repair. Annu. Rev. Biochem. 2021, 90, 107–135. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Brosh, R.M. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Front. Mol. Biosci. 2018, 5, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compe, E.; Egly, J.-M. Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and Beyond. Annu. Rev. Biochem. 2016, 85, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA Double-Strand Breaks and Models of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uliel, L.; Weisman-Shomer, P.; Oren-Jazan, H.; Newcomb, T.; Loeb, L.A.; Fry, M. Human Ku Antigen Tightly Binds and Stabilizes a Tetrahelical Form of the Fragile X Syndrome d(CGG) Expanded Sequence. J. Biol. Chem. 2000, 275, 33134–33141. [Google Scholar] [CrossRef] [Green Version]
- Janel-Bintz, R.; Kuhn, L.; Frit, P.; Chicher, J.; Wagner, J.; Haracska, L.; Hammann, P.; Cordonnier, A.M. Proteomic Analysis of DNA Synthesis on a Structured DNA Template in Human Cellular Extracts: Interplay between NHEJ and Replication-Associated Proteins. Proteomics 2020, 20, 1900184. [Google Scholar] [CrossRef]
- Usdin, K.; House, N.C.M.; Freudenreich, C.H. Repeat Instability during DNA Repair: Insights from Model Systems. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 142–167. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuchs, R.P.; Isogawa, A.; Paulo, J.A.; Fujii, S. Identification of Proteins Specifically Assembled on a Stem-Loop Composed of a CAG Triplet Repeat. DNA 2023, 3, 109-118. https://doi.org/10.3390/dna3020009
Fuchs RP, Isogawa A, Paulo JA, Fujii S. Identification of Proteins Specifically Assembled on a Stem-Loop Composed of a CAG Triplet Repeat. DNA. 2023; 3(2):109-118. https://doi.org/10.3390/dna3020009
Chicago/Turabian StyleFuchs, Robert P., Asako Isogawa, Joao A. Paulo, and Shingo Fujii. 2023. "Identification of Proteins Specifically Assembled on a Stem-Loop Composed of a CAG Triplet Repeat" DNA 3, no. 2: 109-118. https://doi.org/10.3390/dna3020009