Labeling of Polysaccharides with Biotin and Fluorescent Dyes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Na+ to i-Pr2EtNH+ Ion Replacement in Acidic Polysaccharides with the Example of Sodium Hyaluronate 15–30 kDa
2.2.2. Direct Labeling of Acidic Polysaccharides with Fluorescein Isothiocyanate (FITC) with the Example of HA15–30kDa i-Pr2EtNH+ Salt
2.2.3. Synthesis of Dichlorotriazine (DCTA) Reagent, biot-DCTA
2.2.4. Labeling of Polysaccharides with biot-DCTA on the Example of HA15–30kDa Na Salt
2.2.5. Labeling of Bacterial Polysaccharides with DTAF
2.2.6. Fluorescent Labeling of PSs at Carboxyl Groups
Hyaluronic Acids
Xylogalacturonan
2.2.7. Indirect Fluorescent Labeling at Hydroxyl Groups (through the Introduction of the Carboxyl Function)
2.2.8. Probing of Cell Sections with Labeled Polysaccharides
3. Results and Discussion
3.1. FITC Labeling of PSs at Hydroxyl Groups
3.2. Biotinylation of PSs at Hydroxyl Groups
3.3. Fluorescein Labeling of Bacterial PSs Using DTAF Reagent
3.4. Fluorescent Labeling of Acidic Polysaccharide by Carboxyl Groups
3.5. Indirect Fluorescent Labeling at Hydroxyl Groups (through the Introduction of the Caboxyl Function)
3.6. Evaluation of the Label Content
4. Application of the Synthesized Probes, and General Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
- NMR spectra of biot-DCTA


- Stability of Biot-DCTA
- Absorption and fluorescence spectra for the labeled polysaccharides

References
- Balducci, E.; Papi, F.; Capialbi, D.E.; Del Bino, L. Polysaccharides’ Structures and Functions in Biofilm Architecture of Antimicrobial-Resistant (AMR) Pathogens. Int. J. Mol. Sci. 2023, 24, 4030. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, A.V.; Golovchenko, V.V.; Mikshina, P.V.; Patova, O.A.; Gorshkova, T.A.; Bovin, N.V.; Shilova, N.V. Plant Polysaccharide Array for Studying Carbohydrate-Binding Proteins. Biochemistry 2022, 87, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Tuzikov, A.B.; Rapoport, E.M.; Khaidukov, S.V.; Nokel, E.A.; Knirel, Y.A.; Bovin, N.V. Synthesis of Bodipy-Labeled Bacterial Polysaccharides and Their Interaction with Human Dendritic Cells. Glycoconj. J. 2021, 38, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Meunier, F.; Wilkinson, K.J. Nonperturbing Fluorescent Labeling of Polysaccharides. Biomacromolecules 2002, 3, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Oda, Y.; Kinoshita, M.; Masuko, T.; Kakehi, K. Time-Resolved Fluorometric Analysis of Carbohydrates Labeled with Amino-Aromatic Compounds by Reductive Amination. Analyst 2002, 127, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Zhang, X.; Zhou, W.; Huang, L. One-Pot Fluorescent Labeling of Saccharides with Fluorescein-5-Thiosemicarbazide for Imaging Polysaccharides Transported in Living Cells. Carbohydr. Res. 2011, 346, 2156–2164. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Steenvoorden, E.; Koeleman, C.A.M.; Deelder, A.M.; Wuhrer, M. 2-Picoline-Borane: A Non-Toxic Reducing Agent for Oligosaccharide Labeling by Reductive Amination. Proteomics 2010, 10, 2330–2336. [Google Scholar] [CrossRef]
- Cosenza, V.A.; Navarro, D.A.; Stortz, C.A. Usage of α-Picoline Borane for the Reductive Amination of Carbohydrates. Arkivoc 2011, 2011, 182–194. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, J. Preparation and Characterization of the Fluorescent Chitosan Nanoparticle Probe. Chin. J. Anal. Chem. 2006, 34, 1555–1559. [Google Scholar] [CrossRef]
- Moussa, S.H.; Tayel, A.A.; Al-Turki, A.I. Evaluation of Fungal Chitosan as a Biocontrol and Antibacterial Agent Using Fluorescence-Labeling. Int. J. Biol. Macromol. 2013, 54, 204–208. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ichishima, E. Fluorescent Labeling of Sugar-Carboxylate with Water-Soluble Carbodiimide. Biosci. Biotechnol. Biochem. 1992, 56, 186–189. [Google Scholar] [CrossRef]
- Han, Z.R.; Wang, Y.F.; Liu, X.; Wu, J.D.; Cao, H.; Zhao, X.; Chai, W.G.; Yu, G.L. Fluorescent Labeling of Several Glycosaminoglycans and Their Interaction with Anti-Chondroitin Sulfate Antibody. Chin. J. Anal. Chem. 2011, 39, 1352–1357. [Google Scholar] [CrossRef]
- Kunishima, M.; Kawachi, C.; Monta, J.; Terao, K.; Iwasaki, F.; Tani, S. 4-(4,6-Dimethoxy-1,3,5-Triazin-2-Yl)-4-Methyl-Morpholinium Chloride: An Efficient Condensing Agent Leading to the Formation of Amides and Esters. Tetrahedron 1999, 55, 13159–13170. [Google Scholar] [CrossRef]
- D’Este, M.; Eglin, D.; Alini, M. A Systematic Analysis of DMTMM vs EDC/NHS for Ligation of Amines to Hyaluronan in Water. Carbohydr. Polym. 2014, 108, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Bohrn, R.; Potthast, A.; Schiehser, S.; Rosenau, T.; Sixta, H.; Kosma, P. The FDAM Method: Determination of Carboxyl Profiles in Cellulosic Materials by Combining Group-Selective Fluorescence Labelling with GPC. Biomacromolecules 2006, 7, 1743–1750. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, C.; Fackler, K.; Potthast, A. The Fate of 4-O-Methyl Glucuronic Acid in Hardwood Xylan during Alkaline Extraction. ACS Sustain. Chem. Eng. 2017, 5, 1818–1823. [Google Scholar] [CrossRef]
- Abushammala, H.; Mao, J. A Review of the Surface Modification of Cellulose and Nanocellulose Using Aliphatic and Aromatic Mono- and Di-Isocyanates. Molecules 2019, 24, 2782. [Google Scholar] [CrossRef]
- de Belder, A.N.; Granath, K. Preparation and Properties of Fluorescein-Labelled Dextrans. Carbohydr. Res. 1973, 30, 375–378. [Google Scholar] [CrossRef]
- Unnikrishnan, B.S.; Preethi, G.U.; Sreeranganathan, M.; Syama, H.P.; Archana, M.G.; Sreelekha, T.T. Fabrication of Fluorescein Labeled Galactoxyloglucan Polysaccharide for Tumor and Macrophage Tagging. J. Drug Deliv. Sci. Technol. 2019, 52, 863–869. [Google Scholar] [CrossRef]
- Sun, M.; Su, F.; Yang, J.; Gao, Z.; Geng, Y. Fluorescent Labeling of Polysaccharides from Masson Pine Pollen and Its Effect on RAW264.7 Macrophages. Polymers 2018, 10, 372. [Google Scholar] [CrossRef]
- Alasel, M.; Dassinger, N.; Vornicescu, D.; Keusgen, M. Development of a Borreliosis Assay: Mannan Coated Polyethylene Sinter Bodies as a New Platform Technology. Anal. Biochem. 2018, 543, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Vanderah, D. Solubility of N, N′-Disuccinimidyl Carbonate and Its Relevance to Polysaccharide Functionalization. Anal. Biochem. 2021, 626, 114250. [Google Scholar] [CrossRef] [PubMed]
- Heyna, J. Reactive Dyes Containing Vinylsulfonyl Groups. Angew. Chem. Int. Ed. Engl. 1963, 2, 20–23. [Google Scholar] [CrossRef]
- Rapoport, E.M.; Khasbiullina, N.R.; Komarova, V.A.; Ryzhov, I.M.; Gorbatch, M.M.; Tuzikov, A.B.; Khaidukov, S.V.; Popova, I.S.; Korchagina, E.Y.; Henry, S.M.; et al. Localization of Synthetic Glycolipids in the Cell and the Dynamics of Their Insertion/Loss. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183645. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, K.A.; Mena, J.A.; Male, K.B.; Hrapovic, S.; Kamen, A.; Luong, J.H.T. Effect of Surface Charge on the Cellular Uptake and Cytotoxicity of Fluorescent Labeled Cellulose Nanocrystals. ACS Appl. Mater. Interfaces 2010, 2, 2924–2932. [Google Scholar] [CrossRef]
- Hess, R.; Pearse, A.G.E. Labelling of Proteins with Cellulose-Reactive Dyes. Nature 1959, 183, 260–261. [Google Scholar] [CrossRef]
- Abitbol, T.; Palermo, A.; Moran-Mirabal, J.M.; Cranston, E.D. Fluorescent Labeling and Characterization of Cellulose Nanocrystals with Varying Charge Contents. Biomacromolecules 2013, 14, 3278–3284. [Google Scholar] [CrossRef]
- Petrova, A.; Sibgatullina, G.; Gorshkova, T.; Kozlova, L. Dynamics of Cell Wall Polysaccharides during the Elongation Growth of Rye Primary Roots. Planta 2022, 255, 108. [Google Scholar] [CrossRef]
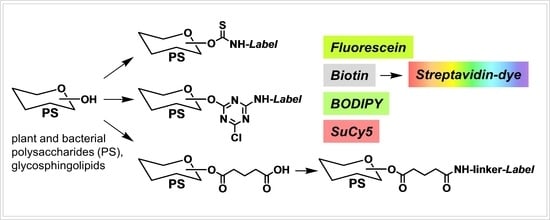
 ) and biotin tags. Reagents and conditions: (A) PS in the form of i-Pr2EtN salt in DMSO, FITC, i-Pr2EtN, 40 h/50 °C; (B) native PS, biot-DCTA, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
) and biotin tags. Reagents and conditions: (A) PS in the form of i-Pr2EtN salt in DMSO, FITC, i-Pr2EtN, 40 h/50 °C; (B) native PS, biot-DCTA, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
 ) and biotin tags. Reagents and conditions: (A) PS in the form of i-Pr2EtN salt in DMSO, FITC, i-Pr2EtN, 40 h/50 °C; (B) native PS, biot-DCTA, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
) and biotin tags. Reagents and conditions: (A) PS in the form of i-Pr2EtN salt in DMSO, FITC, i-Pr2EtN, 40 h/50 °C; (B) native PS, biot-DCTA, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
 ). (A) Capsular polysaccharide (CPS Acinetobacter baumannii NIPH60 K43). (B) Polysaccharide with O-antigen (O-PS Escherichia coli O41), containing an amino group in the CORE (inner core part of the source LPS from which the PS was obtained). Reagents and conditions for (A,B): DTAF, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
). (A) Capsular polysaccharide (CPS Acinetobacter baumannii NIPH60 K43). (B) Polysaccharide with O-antigen (O-PS Escherichia coli O41), containing an amino group in the CORE (inner core part of the source LPS from which the PS was obtained). Reagents and conditions for (A,B): DTAF, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
 ). (A) Capsular polysaccharide (CPS Acinetobacter baumannii NIPH60 K43). (B) Polysaccharide with O-antigen (O-PS Escherichia coli O41), containing an amino group in the CORE (inner core part of the source LPS from which the PS was obtained). Reagents and conditions for (A,B): DTAF, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
). (A) Capsular polysaccharide (CPS Acinetobacter baumannii NIPH60 K43). (B) Polysaccharide with O-antigen (O-PS Escherichia coli O41), containing an amino group in the CORE (inner core part of the source LPS from which the PS was obtained). Reagents and conditions for (A,B): DTAF, water/DMSO 1:1, i-Pr2EtN (pH~9), 4 h/20 °C.
 )-labeled hyaluronic acid (15–30 kDa), reagents and conditions: sodium hyaluronate, BODIPY amine, HONSu, EDAC, water/DMF, 7 h/20° C. (B) SuCy5-labeled (
)-labeled hyaluronic acid (15–30 kDa), reagents and conditions: sodium hyaluronate, BODIPY amine, HONSu, EDAC, water/DMF, 7 h/20° C. (B) SuCy5-labeled ( ) xylogalacturonan from baobab fruit Adansonia digitata L., reagents and conditions: i-Pr2EtNH+ salt of xylogalacturonan, SuCy5 amine, HOBT, DCC, DMF, 24 h/25 °C.
) xylogalacturonan from baobab fruit Adansonia digitata L., reagents and conditions: i-Pr2EtNH+ salt of xylogalacturonan, SuCy5 amine, HOBT, DCC, DMF, 24 h/25 °C.
 )-labeled hyaluronic acid (15–30 kDa), reagents and conditions: sodium hyaluronate, BODIPY amine, HONSu, EDAC, water/DMF, 7 h/20° C. (B) SuCy5-labeled (
)-labeled hyaluronic acid (15–30 kDa), reagents and conditions: sodium hyaluronate, BODIPY amine, HONSu, EDAC, water/DMF, 7 h/20° C. (B) SuCy5-labeled ( ) xylogalacturonan from baobab fruit Adansonia digitata L., reagents and conditions: i-Pr2EtNH+ salt of xylogalacturonan, SuCy5 amine, HOBT, DCC, DMF, 24 h/25 °C.
) xylogalacturonan from baobab fruit Adansonia digitata L., reagents and conditions: i-Pr2EtNH+ salt of xylogalacturonan, SuCy5 amine, HOBT, DCC, DMF, 24 h/25 °C.
 ) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C; iii, 2-propanol/MeOH/water, Et3N, 2 h/50 °C; iv, SuCy5 (
) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C; iii, 2-propanol/MeOH/water, Et3N, 2 h/50 °C; iv, SuCy5 ( ) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C. For details see Section 2.2.7.
) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C. For details see Section 2.2.7.
 ) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C; iii, 2-propanol/MeOH/water, Et3N, 2 h/50 °C; iv, SuCy5 (
) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C; iii, 2-propanol/MeOH/water, Et3N, 2 h/50 °C; iv, SuCy5 ( ) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C. For details see Section 2.2.7.
) amine, DCC, HOBt, i-Pr2EtN, DMF, 24 h/37 °C. For details see Section 2.2.7.

| # | Structure | Source | Type |
|---|---|---|---|
| 1 | Hyaluronic acid 8–15 kDa -4GlcAβ1-3GlcNAcβ1- | Streptococcus equi, zooepidemicus Contipro Biotech (Czech Republic) | Glycosaminoglycan (GAG) |
| 2 | Hyaluronic acid 15–30 kDa -4GlcAβ1-3GlcNAcβ1- | Streptococcus equi, zooepidemicus Contipro Biotech (Czech Republic) | GAG |
| 3 | Xylogalacturonan | Adansonia digitata L., baobab fruit | pectin |
| # | Structure | Source | Type |
|---|---|---|---|
| 4 | Hyaluronic acid 15–30 kDa -4GlcAβ1-3GlcNAcβ1- | Streptococcus equi, zooepidemicus Contipro Biotech (Czech Republic) | GAG |
| 5 | Homogalacturonan predominance | Malus sp., apple, Fluka76282 | pectin |
| 6 | Ara predominance | Beta vulgaris L., sugar beet, Megazyme 11078-27-6 | pectin |
| 7 | Ara and Gal predominance | Larix sp. larch, Megazyme 9036-66-2 | pectin |
| 8 | Ara and Gal predominance | Acacia sp. acacia, Sigma G9752 | pectin |
| 9 | Xylogalacturonan * | Adansonia digitata L., baobab fruit | pectin |
| # | Structure * | Source | Type ** |
|---|---|---|---|
| 1 | -2Glcβ1-6GlcNAcα1-3FucNAcα1-GlcNAcβ1- | Escherichia coli O12 | O-PS |
| 2 | Galα1-2Galα1-2(Galβ1-4)Glcα1-3Glcα1-/inner core-lipid A/ | Escherichia coli O14 | LPS(R) |
| 3 | -3Galα1-3(GlcAβ1-4)Fucα1-4GlcNAcβ1-3Fucα1-3GlcNAcβ1- | Escherichia coli O41 | O-PS |
| 4 | -4GalAα1-2Rhaα1-2Ribfβ1-4Galβ1-3GlcNAcβ1- | Escherichia coli O54 | O-PS |
| 5 | -5(Glcα1-2)Galfβ1-5(Glcα1-2)Galfβ1-3Galfβ1- | Escherichia coli O62 | O-PS |
| 6 | -4(Aci5Ac7Acα)Galα1-3FucNAcα1-3FucNAcα1- | Acinetobacter baumannii UMB001 K13 | CPS |
| 7 | -3GalNAcA4Acα1-4GalNAcA3(%)Acα1-4GalNAcA3(%)Acα1-3QuiNAc4NAcβ1- | Acinetobacter baumannii LUH5535 K35 | CPS |
| 8 | -3(Pse5Ac7RHbα2-4)Ribβ1-3GalNAcβ1- | Acinetobacter baumannii LUH5550 K42 | CPS |
| 9 | -6GlcNAcα1-4(Glcβ1-3)GalNAcα1-3GlcNAcα1- | Acinetobacter baumannii NIPH60 K43 | CPS |
| 10 | -4ManA2Acβ1-4(GalNAcα1-3)FucNAcα1-3DFucNAcα1- | Acinetobacter baumannii MAR13-1452 K125 | CPS |
| 11 | -2Rha3Acα1-2Rhaα1-4GalAβ1-3GalNAcβ1- | Shigella flexneri type 6 (5-F6 51579) | O-PS |
| 12 | -2Rhaα1-2Rhaα1-3Rhaα1-3GlcNAcβ1- | Shigella flexneri type Y (51581 Y) | O-PS |
| 13 | -2(EtN1-P-3)Rhaα1-2(EtN1-P-3)Rhaα1-3Rhaα1-3GlcNAcβ1- | Shigella flexneri Yv (Yv) | O-PS |
| 14 | -2Rha3Acα1-2Rhaα1-3(Glcα1-4)Rhaα1-3GlcNAc6Acβ1- | Shigella flexneri 2a2-2 (2a) | O-PS |
| 15 | -3GlcNAcβ1-2Rha3Acα1-2(Glcα1-3)Rhaα1-3Rhaα1- | Shigella flexneri type 5a1 (F-5a) | O-PS |
| 16 | -2Rhaα1-2Rhaα1-2Rhaα1-2Glcα1-3GlcNAc6Acα1- | Escherichia coli O19ab | O-PS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuzikov, A.; Shilova, N.; Ovchinnikova, T.; Nokel, A.; Patova, O.; Knirel, Y.; Chernova, T.; Gorshkova, T.; Bovin, N. Labeling of Polysaccharides with Biotin and Fluorescent Dyes. Polysaccharides 2024, 5, 1-15. https://doi.org/10.3390/polysaccharides5010001
Tuzikov A, Shilova N, Ovchinnikova T, Nokel A, Patova O, Knirel Y, Chernova T, Gorshkova T, Bovin N. Labeling of Polysaccharides with Biotin and Fluorescent Dyes. Polysaccharides. 2024; 5(1):1-15. https://doi.org/10.3390/polysaccharides5010001
Chicago/Turabian StyleTuzikov, Alexander, Nadezhda Shilova, Tatiana Ovchinnikova, Alexey Nokel, Olga Patova, Yuriy Knirel, Tatiana Chernova, Tatiana Gorshkova, and Nicolai Bovin. 2024. "Labeling of Polysaccharides with Biotin and Fluorescent Dyes" Polysaccharides 5, no. 1: 1-15. https://doi.org/10.3390/polysaccharides5010001