Photocatalytic Duplex-Based DNAzymes Switched by an Abasic Site
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
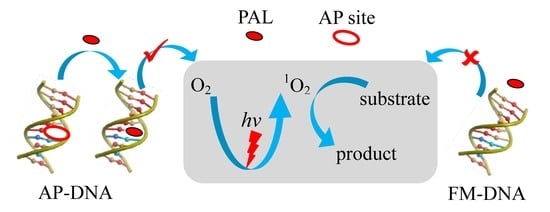
3.1. Duplex-Based DNAzyme with the AP Site as the Active Site
3.2. Optimized Conditions of DNAzyme
3.3. Mechanism of the DNAzyme
3.4. Stability of the DNAzyme
3.5. Switch of the DNAzyme Activity Using Cascade UDG
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ouyang, Y.; O’Hagan, M.P.; Willner, B.; Willner, I. Aptamer-Modified Homogeneous Catalysts, Heterogenous Nanoparticle Catalysts, and Photocatalysts: Functional “Nucleoapzymes”, “Aptananozymes”, and “Photoaptazymes”. Adv. Mater. 2023, 2023, 2210885. [Google Scholar] [CrossRef]
- Lan, T.; Lu, Y. Metal Ion-Dependent DNAzymes and Their Applications as Biosensors. Met. Ions Life Sci. 2012, 10, 217–248. [Google Scholar] [PubMed]
- Yu, Y.; Zhang, Q.; Gao, H.; Yan, C.; Zheng, X.; Yang, T.; Zhou, X.; Shao, Y. Metalloenzyme-Mimic Innate G-quadruplex DNAzymes Using Directly Coordinated Metal Ions as Active Centers. Dalton Trans. 2020, 49, 13160–13166. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Vazin, M.; Yu, T.; Ding, J.; Liu, J. In Vitro Selection of Chromium-Dependent DNAzymes for Sensing Chromium(III) and Chromium(VI). Chem. Eur. J. 2016, 22, 9835–9840. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. Mechanism and Utility of an RNA-Cleaving DNA Enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A General-Purpose RNA-Cleaving DNA Enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Li, T.; Li, B.; Wang, E. Potassium-Sensitive G-Quadruplex DNA for Sensitive Visible Potassium Detection. Analyst 2010, 135, 71–75. [Google Scholar] [CrossRef]
- Nakayama, S.; Sintim, H.O. Colorimetric Split G-Quadruplex Probes for Nucleic Acid Sensing: Improving Reconstituted DNAzyme’s Catalytic Efficiency via Probe Remodeling. J. Am. Chem. Soc. 2009, 131, 10320–10333. [Google Scholar] [CrossRef]
- Elbaz, J.; Shlyahovsky, B.; Willner, I. A DNAzyme Cascade for the Amplified Detection of Pb2+ Ions or L-Histidine. Chem. Commun. 2008, 13, 1569–1571. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Potassium-Lead-Switched G-Quadruplexes: A New Class of DNA Logic Gates. J. Am. Chem. Soc. 2009, 131, 15082–15083. [Google Scholar] [CrossRef]
- Boiteux, S.; Guillet, M. Abasic Sites in DNA: Repair and Biological Consequences in Saccharomyces Cerevisiae. DNA Repair 2004, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Y.; Wu, T.; Zhang, L.; Liu, H.; Zhou, X.; Shao, Y. Recognition of DNA Abasic Site Nanocavity by FluoroPhore-Switched Probe: Suitable for All Sequence Environments. Spectrochim. Acta Part A 2016, 153, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Nishizawa, S.; Minagawa, M.; Teramae, N. Use of Abasic Site-Containing DNA Strands for Nucleobase Recognition in Water. J. Am. Chem. Soc. 2003, 125, 8982–8983. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Zhao, C.; Rajendar, B.; Thiagarajan, V.; Sato, Y.; Nishizawa, S.; Teramae, N. Effect of the Bases Flanking an Abasic Site on the Recognition of Nucleobase by Amiloride. Biochim. Biophys. Acta 2010, 1800, 599–610. [Google Scholar] [CrossRef]
- Wu, F.; Sun, Y.; Shao, Y.; Xu, S.; Liu, G.; Peng, J.; Liu, L. DNA Abasic Site-Selective Enhancement of Sanguinarine Fluorescence with a Large Emission Shift. PLoS ONE 2012, 7, e48251. [Google Scholar] [CrossRef] [Green Version]
- Dougherty, T.J.; Gomer, C.J.; Henderson, B.W.; Jori, G.; Kessel, D.; Korbelik, M.; Moan, J.; Peng, Q. Photodynamic Therapy. J. Natl. Cancer Inst. 1998, 90, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Ethirajan, M.; Chen, Y.; Joshi, P.; Pandey, R.K. The Role of Porphyrin Chemistry in Tumor Imaging and Photodynamic Therapy. Chem. Soc. Rev. 2011, 40, 340–362. [Google Scholar] [CrossRef]
- Rotaru, A.; Mokhir, A. Nucleic Acid Binders Activated by Light of Selectable Wavelength. Angew. Chem. Int. Ed. 2007, 46, 6180–6183. [Google Scholar] [CrossRef]
- Tørring, T.; Helmig, S.; Ogilby, P.R.; Gothelf, K.V. Singlet Oxygen in DNA Nanotechnology. Acc. Chem. Res. 2014, 47, 1799–1806. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Dong, Z.; Hu, H.; Yang, Q.; Hou, X.; Wu, P. DNA-Modulated Photosensitization: Current Status and Future Aspects in Biosensing and Environmental Monitoring. Anal. Bioanal. Chem. 2019, 411, 4415–4423. [Google Scholar] [CrossRef]
- Tardivo, J.P.; Del Giglio, A.; de Oliveira, C.S.; Gabrielli, D.S.; Junqueira, H.C.; Tada, D.B.; Severino, D.; de Fátima Turchiello, R.; Baptista, M.S. Methylene Blue in Photodynamic Therapy: From Basic Mechanisms to Clinical Applications. Photodiagn. Photodyn. Ther. 2005, 2, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Thivierge, C.; Nowak-Sliwinska, P.; Han, J.; van den Bergh, H.; Wagnières, G.; Burgess, K.; Lee, H.B. In Vitro and in vivo Photocytotoxicity of Boron Dipyrromethene Derivatives for Photodynamic Therapy. J. Med. Chem. 2010, 53, 2865–2874. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Li, Z.; Liu, Z.; Ren, J.; Yang, X.; Qu, X. Photosensitizer Incorporated G-Quadruplex DNA-Functionalized Magnetofluorescent Nanoparticles for Targeted Magnetic Resonance/Fluorescence Multimodal Imaging and Subsequent Photodynamic Therapy of Cancer. Chem. Commun. 2012, 48, 6556–6558. [Google Scholar] [CrossRef]
- Yuan, Q.; Wu, Y.; Wang, J.; Lu, D.; Zhao, Z.; Liu, T.; Zhang, X.; Tan, W. Targeted Bioimaging and Photodynamic Therapy Nanoplatform Using an Aptamer-Guided G-Quadruplex DNA Carrier and Near-Infrared Light. Angew. Chem. Int. Ed. 2013, 52, 13965–13969. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A Hitchhiker’s Guide to G-Quadruplex Ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Liu, Y.C.; Liang, H. Water-soluble Ruthenium (II) Complexes with Chiral 4-(2,3-Dihydroxypropyl)-Formamide Oxoaporphine (FOA): In Vitro and in vivo Anticancer Activity by Stabilization of G-Quadruplex DNA, Inhibition of Telomerase Activity, and Induction of Tumor Cell Apoptosis. J. Med. Chem. 2015, 58, 4771–4789. [Google Scholar]
- Gao, L.; Tong, X.; Ye, T.; Gao, H.; Zhang, Q.; Yan, C.; Yu, Y.; Fei, Y.; Zhou, X.; Shao, Y. G-Quadruplex-Based Photooxidase Driven by Visible Light. ChenCatChem 2020, 12, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, K.; Hirano, T. The Microenvironment of DNA Switches the Activity of Singlet Oxygen Generation Photosensitized by Berberine and Palmatine. Photochem. Photobiol. 2008, 84, 202–208. [Google Scholar] [CrossRef]
- Hirakawa, K.; Hirano, T.; Nishimura, Y.; Arai, T.; Nosaka, Y. Dynamics of Singlet Oxygen Generation by DNA-Binding Photosensitizers. J. Phys. Chem. B 2012, 116, 3037–3044. [Google Scholar] [CrossRef]
- Hirakawa, K.; Nishimura, Y.; Arai, T.; Okazaki, S. Singlet Oxygen Generating Activity of an Electron Donor Connecting Porphyrin Photosensitizer Can Be Controlled by DNA. J. Phys. Chem. B 2013, 117, 13490–13496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, C.; Xu, S.; Chen, J.; Zeng, Y.; Wu, P.; Hou, X. Photocatalytic Oxidation of TMB with The Double Stranded DNA-SYBR Green I Complex for Label-Free and Universal Colorimetric Bioassay. Chem. Commun. 2015, 51, 14465–14468. [Google Scholar] [CrossRef] [PubMed]
- Stefan, L.; Denat, F.; Monchaud, D. Insights into How Nucleotide Supplements Enhance the Peroxidase-Mimicking DNAzyme Activity of the G-Quadruplex/Hemin System. Nucleic Acids Res. 2012, 40, 8759–8772. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Zhang, J.; Ding, Y.; Zhang, X.; Xu, K.; Hou, X.; Wu, P. Modulation of the Singlet Oxygen Generation from the Double Strand DNA-SYBR Green I Complex Mediated by T-Melamine-T Mismatch for Visual Detection of Melamine. Anal. Chem. 2017, 89, 5101–5106. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Fettinger, J.C.; Davis, J.T. Homochiral G-quadruplexes with Ba2+ but Not with K+: The Cation Programs Enantiomeric Self-Recognition. J. Am. Chem. Soc. 2001, 123, 6738–6739. [Google Scholar] [CrossRef]






| Name | Sequence | Remark |
|---|---|---|
| AXA-A | 5′-ATGGTGAXAGCAGCG-3′ 3′-TACCACTATCGTCGC-5′ | X = AP site |
| AXA-C | 5′-ATGGTGAXAGCAGCG-3′ 3′-TACCACTCTCGTCGC-5′ | X = AP site |
| AXA-G | 5′-ATGGTGAXAGCAGCG-3′ 3′-TACCACTGTCGTCGC-5′ | X = AP site |
| AXA-T | 5′-ATGGTGAXAGCAGCG-3′ 3′-TACCACTTTCGTCGC-5′ | X = AP site |
| FM-DNA | 5′-ATGGTGATAGCAGCG-3′ 3′-TACCACTATCGTCGC-5′ | |
| AUA-A | 5′-ATGGTGAUAGCAGCG-3′ 3′-TACCACTATCGTCGC-5′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Tian, R.; Shao, Y. Photocatalytic Duplex-Based DNAzymes Switched by an Abasic Site. Chemistry 2023, 5, 1497-1507. https://doi.org/10.3390/chemistry5030102
Gao L, Tian R, Shao Y. Photocatalytic Duplex-Based DNAzymes Switched by an Abasic Site. Chemistry. 2023; 5(3):1497-1507. https://doi.org/10.3390/chemistry5030102
Chicago/Turabian StyleGao, Longlong, Rui Tian, and Yong Shao. 2023. "Photocatalytic Duplex-Based DNAzymes Switched by an Abasic Site" Chemistry 5, no. 3: 1497-1507. https://doi.org/10.3390/chemistry5030102