Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Thiol Modification of DeepTipTM Probes and MicroDeckTM Substrates
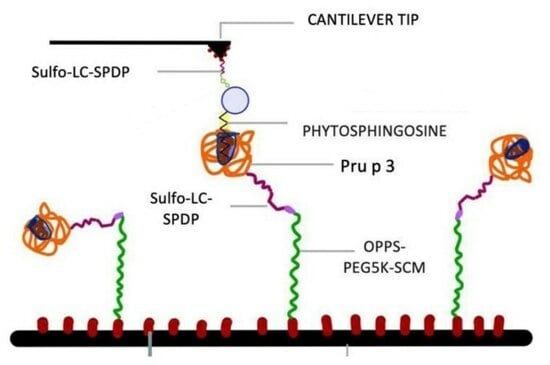
2.3. Decoration of the DeepTipTM Probes with Phytosphingosine
2.4. Modification of Pru p 3 with Sulfo-LC-SPDP and Decoration of the MicroDeckTM Substrates
2.5. Determination of the Pru p 3-phytosphingosine Force–Distance (F-d) Curves
3. Results and Discussion
3.1. Classification of the Force–Distance (F-d) Curves
3.2. Interpretation of the Force–Distance (F-d) Curves
3.3. New Perspectives Opened by High-Yield A-AFM
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmitt, O.H. Some interesting and useful biomimetic transforms. In Proceedings of the Third International Biophysics Congress, Cambridge, MA, USA, 29 August–3 September 1969. [Google Scholar]
- Mirvakili, S.M.; Hunter, I.W. Artificial Muscles: Mechanisms, Applications, and Challenges. Adv. Mater. 2018, 30, 1704407. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.R.M.; Das, S.; Kaufman, Y.; Israelachvili, J.N.; Waite, J.H. Interfacial pH during mussel adhesive plaque formation. Biofouling 2015, 31, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Badin, R.; Burgain, J.; Desobry, S.; Bhandari, B.; Prakash, S.; Gaiani, C. Probing maltodextrins surface properties by atomic force microscopy: Interplay of glass transition and reconstitution properties. Food Hydrocoll. 2022, 132, 107853. [Google Scholar] [CrossRef]
- Horber, J.K.H.; Miles, M.J. Scanning Probe Evolution in Biology. Science 2003, 302, 1002–1005. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Dumitru, A.C.; Conrard, L.; Giudice, C.L.; Henriet, P.; Veiga-Da-Cunha, M.; Derclaye, S.; Tyteca, D.; Alsteens, D. High-resolution mapping and recognition of lipid domains using AFM with toxin-derivatized probes. Chem. Commun. 2018, 54, 6903–6906. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends Cell Biol. 2011, 21, 461–469. [Google Scholar] [CrossRef]
- Daza, R.; Cruces, J.; Arroyo-Hernández, M.; Marí-Buyé, N.; De la Fuente, M.; Plaza, G.R.; Elices, M.; Pérez-Rigueiro, J.; Guinea, G.V. Topographical and mechanical characterization of living eukaryotic cells on opaque substrates: Development of a general procedure and its application to the study of non-adherent lymphocytes. Phys. Biol. 2015, 12, 026005. [Google Scholar] [CrossRef]
- Frisbie, C.D.; Rozsnyai, L.F.; Noy, A.; Wrighton, M.S.; Lieber, C.M. Functional Group Imaging by Chemical Force Microscopy. Science 1994, 265, 2071–2074. [Google Scholar] [CrossRef]
- Dammer, U.; Hegner, M.; Anselmetti, D.; Wagner, P.; Dreier, M.; Huber, W.; Güntherodt, H. Specific antigen/antibody interactions measured by force microscopy. Biophys. J. 1996, 70, 2437–2441. [Google Scholar] [CrossRef] [PubMed]
- Riener, C.K.; Stroh, C.M.; Ebner, A.; Klampfl, C.; Gall, A.A.; Romanin, C.; Lyubchenko, Y.L.; Hinterdorfer, P.; Gruber, H.J. Simple test system for single molecule recognition force microscopy. Anal. Chim. Acta 2003, 479, 59–75. [Google Scholar] [CrossRef]
- Kienberger, F.; Ebner, A.; Gruber, H.J.; Hinterdorfer, P. Molecular Recognition Imaging and Force Spectroscopy of Single Biomolecules. Accounts Chem. Res. 2005, 39, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Siewny, M.G.W.; Edwards, D.T.; Sanders, A.W.; Perkins, T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 2017, 355, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Grandbois, M.; Dettmann, W.; Benoit, M.; Gaub, H.E. Affinity Imaging of Red Blood Cells Using an Atomic Force Microscope. J. Histochem. Cytochem. 2000, 48, 719–724. [Google Scholar] [CrossRef]
- Dumitru, A.C.; Herruzo, E.T.; Rausell, E.; Ceña, V.; Garcia, R. Unbinding forces and energies between a siRNA molecule and a dendrimer measured by force spectroscopy. Nanoscale 2015, 7, 20267–20276. [Google Scholar] [CrossRef]
- Barattin, R.; Voyer, N. Chemical modifications of AFM tips for the study of molecular recognition events. Chem. Commun. 2008, 13, 1513–1532. [Google Scholar] [CrossRef]
- Volcke, C.; Gandhiraman, R.P.; Gubala, V.; Doyle, C.; Fonder, G.; Thiry, P.A.; Cafolla, A.A.; James, B.; Williams, D.E. Plasma functionalization of AFM tips for measurement of chemical interactions. J. Colloid Interface Sci. 2010, 348, 322–328. [Google Scholar] [CrossRef]
- Bergkvist, M.; Cady, N.C. Chemical Functionalization and Bioconjugation Strategies for Atomic Force Microscope Cantilevers. In Bioconjugation Protocols: Strategies and Methods; Mark, S.S., Ed.; Humana Press: Totowa, NJ, USA, 2011; Volume 751, pp. 381–400. [Google Scholar] [CrossRef]
- Wildling, L.; Unterauer, B.; Zhu, R.; Rupprecht, A.; Haselgrübler, T.; Rankl, C.; Ebner, A.; Vater, D.; Pollheimer, P.; Pohl, E.E.; et al. Linking of Sensor Molecules with Amino Groups to Amino-Functionalized AFM Tips. Bioconjug. Chem. 2011, 22, 1239–1248. [Google Scholar] [CrossRef]
- Ebner, A.; Hinterdorfer, P.; Gruber, H.J. Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy 2007, 107, 922–927. [Google Scholar] [CrossRef]
- Bioactive Surfaces S.L. Available online: www.bioactivesurfaces.com (accessed on 1 September 2023).
- Corregidor, D.; Tabraue, R.; Colchero, L.; Daza, R.; Elices, M.; Guinea, G.V.; Pérez-Rigueiro, J. High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes. Molecules 2023, 28, 226. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Ray, C.; Kirkpatrick, A.; Lad, N.; Akhremitchev, B.B. Effects of Multiple-Bond Ruptures on Kinetic Parameters Extracted from Force Spectroscopy Measurements: Revisiting Biotin-Streptavidin Interactions. Biophys. J. 2008, 95, 3964–3976. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, S.M.; Schendel, L.C.; Gaub, H.E.; Bernardi, R.C. Streptavidin/biotin: Tethering geometry defines unbinding mechanics. Sci. Adv. 2020, 6, eaay5999. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Chilkoti, A.; Moy, V.T. Direct force measurements of the streptavidin–biotin interaction. Biomol. Eng. 1999, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Chen, A.; Kolb, P.; Moy, V.T. Energy Landscape of Streptavidin−Biotin Complexes Measured by Atomic Force Microscopy. Biochemistry 2000, 39, 10219–10223. [Google Scholar] [CrossRef]
- Teulon, J.-M.; Delcuze, Y.; Odorico, M.; Chen, S.-w.W.; Parot, P.; Pellequer, J.-L. Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 2011, 24, 490–502. [Google Scholar] [CrossRef]
- Missaoui, K.; Gonzalez-Klein, Z.; Pazos-Castro, D.; Hernandez-Ramirez, G.; Garrido-Arandia, M.; Brini, F.; Diaz-Perales, A.; Tome-Amat, J. Plant non-specific lipid transfer proteins: An overview. Plant Physiol. Biochem. 2022, 171, 115–127. [Google Scholar] [CrossRef]
- Gonzalez-Klein, Z.; Cuevas-Zuviria, B.; Wangorsch, A.; Hernandez-Ramirez, G.; Pazos-Castro, D.; Oeo-Santos, C.; Romero-Sahagun, A.; Pacios, L.F.; Tome-Amat, J.; Scheurer, S.; et al. The key to the allergenicity of lipid transfer protein (LTP) ligands: A structural characterization. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158928. [Google Scholar] [CrossRef]
- Cubells-Baeza, N.; Gómez-Casado, C.; Tordesillas, L.; Ramírez-Castillejo, C.; Garrido-Arandia, M.; González-Melendi, P.; Herrero, M.; Pacios, L.F.; Díaz-Perales, A. Identification of the ligand of Pru p 3, a peach LTP. Plant Mol. Biol. 2017, 94, 33–44. [Google Scholar] [CrossRef]
- Israelachvili, J.N. Intermolecular and Surface Forces, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Pazos-Castro, D.; Gonzalez-Klein, Z.; Montalvo, A.Y.; Hernandez-Ramirez, G.; Romero-Sahagun, A.; Esteban, V.; Garrido-Arandia, M.; Tome-Amat, J.; Diaz-Perales, A. NLRP3 priming due to skin damage precedes LTP allergic sensitization in a mouse model. Sci. Rep. 2022, 12, 3329. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef]
- Pérez-Rigueiro, J. Biological Materials and Biomaterials; UPM Press: Madrid, Spain, 2023; Chapter 24. [Google Scholar]
- Yang, B.; Liu, Z.; Liu, H.; Nash, M.A. Next Generation Methods for Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes. Front. Mol. Biosci. 2020, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S. Statistical and Computational Methods in Data Analysis, 3rd ed.; Springer: New York, NY, USA, 1998. [Google Scholar]
- Pasquato, N.; Berni, R.; Folli, C.; Folloni, S.; Cianci, M.; Pantano, S.; Helliwell, J.R.; Zanotti, G. Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens. J. Mol. Biol. 2006, 356, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Zuviría, B.; Garrido-Arandia, M.; Díaz-Perales, A.; Pacios, L.F. Energy Landscapes of Ligand Motion Inside the Tunnel-Like Cavity of Lipid Transfer Proteins: The Case of the Pru p 3 Allergen. Int. J. Mol. Sci. 2019, 20, 1432. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Carreras, D.; Gavilan-Herrera, M.; Martinez-Martin, I.; Suay-Corredera, C.; Dumitru, A.C.; Galán, E.H.; Alegre-Cebollada, J. Towards a new modular polyprotein system compatible with single-molecule force spectroscopy by atomic force microscopy and magnetic tweezers. Biophys. J. 2023, 122, 304a. [Google Scholar] [CrossRef]
- Milles, L.F.; Schulten, K.; Gaub, H.E.; Bernardi, R.C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 2018, 359, 1527–1533. [Google Scholar] [CrossRef]
- Rico, F.; Russek, A.; González, L.; Grubmüller, H.; Scheuring, S. Heterogeneous and rate-dependent streptavidin–biotin unbinding revealed by high-speed force spectroscopy and atomistic simulations. Proc. Natl. Acad. Sci. USA 2019, 116, 6594–6601. [Google Scholar] [CrossRef]
- Verma, P. Tip-Enhanced Raman Spectroscopy: Technique and Recent Advances. Chem. Rev. 2017, 117, 6447–6466. [Google Scholar] [CrossRef]
- Zhang, Z.; Sheng, S.; Wang, R.; Sun, M. Tip-Enhanced Raman Spectroscopy. Anal. Chem. 2016, 88, 9328–9346. [Google Scholar] [CrossRef]





| Curve | X2 | χ20.05 (f = 9) |
|---|---|---|
| Single elastomeric | 12.2 | 15.51 |
| First peak —two-peak elastomeric | 13.2 | |
| Second peak —two-peak elastomeric | 2.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daza, R.; Garrido-Arandia, M.; Corregidor-Ortiz, D.; Pérez, C.I.; Colchero, L.; Tabraue-Rubio, R.; Elices, M.; Guinea, G.V.; Diaz-Perales, A.; Pérez-Rigueiro, J. Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System. Biomimetics 2023, 8, 595. https://doi.org/10.3390/biomimetics8080595
Daza R, Garrido-Arandia M, Corregidor-Ortiz D, Pérez CI, Colchero L, Tabraue-Rubio R, Elices M, Guinea GV, Diaz-Perales A, Pérez-Rigueiro J. Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System. Biomimetics. 2023; 8(8):595. https://doi.org/10.3390/biomimetics8080595
Chicago/Turabian StyleDaza, Rafael, María Garrido-Arandia, Daniel Corregidor-Ortiz, Carla Isabel Pérez, Luis Colchero, Raquel Tabraue-Rubio, Manuel Elices, Gustavo V. Guinea, Araceli Diaz-Perales, and José Pérez-Rigueiro. 2023. "Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System" Biomimetics 8, no. 8: 595. https://doi.org/10.3390/biomimetics8080595