NTH2 1271_1272delTA Gene Disruption Results in Salt Tolerance in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Strain, Medium, and Growth Conditions
2.2. Preparation of the CRISPR Plasmid
2.3. S. cerevisiae Competent Cell Preparation and Transformation
2.4. Selection and Sequencing Confirmation of Mutants
2.5. nth1 893_894insT and nth2 1271_1272delTA Strain Phenotypes
2.6. Statistical Software
2.7. Scanning Electron Microscopy (SEM)
2.8. Transmission Electron Microscopy (TEM)
3. Results
3.1. Confirmation of NTH1-NTH2 Gene-Mutated Cells
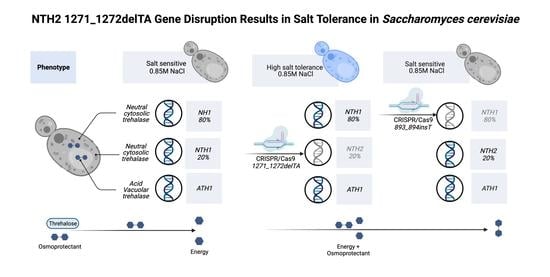
3.2. Behavior of the nth2 1271_1272delTA Strain under Salinity Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Zhang, Y.; Li, H. Regulation of trehalose, a typical stress protectant, on central metabolisms, cell growth and division of Saccharomyces cerevisiae CEN.PK113-7D. Food Microbiol. 2020, 89, 103459. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Gonzalez-Uriarte, A.; Griffiths, C.A.; Hassani-Pak, K. The Role of Trehalose 6-Phosphate in Crop Yield and Resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Fichtner, F.; Lunn, J.E. The Role of Trehalose 6-Phosphate (Tre6P) in Plant Metabolism and Development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Seitl, I.; Mu, W.; Zhang, T.; Stressler, T.; Fischer, L.; Jiang, B. Biotechnical production of trehalose through the trehalose synthase pathway: Current status and future prospects. Appl. Microbiol. Biotechnol. 2018, 102, 2965–2976. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, A.; Golovina, E.A.; Gervais, P.; Dupont, S.; Beney, L. Anhydrobiosis: Inside yeast cells. Biotechnol. Adv. 2019, 37, 51–67. [Google Scholar] [CrossRef]
- Glatz, A.; Pilbat, A.-M.; Németh, G.L.; Vince-Kontár, K.; Jósvay, K.; Hunya, Á.; Udvardy, A.; Gombos, I.; Péter, M.; Balogh, G.; et al. Involvement of small heat shock proteins, trehalose, and lipids in the thermal stress management in Schizosaccharomyces pombe. Cell Stress Chaperon 2016, 21, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Chen, D.; Wang, G.; Zhang, C.; Du, L.; Liu, S.; Zhao, Y.; Xiao, D. Improving freeze-tolerance of baker’s yeast through seamless gene deletion of NTH1 and PUT1. J. Ind. Microbiol. Biotechnol. 2016, 43, 817–828. [Google Scholar] [CrossRef]
- Tapia, H.; Koshland, D.E. Trehalose Is a Versatile and Long-Lived Chaperone for Desiccation Tolerance. Curr. Biol. 2014, 24, 2758–2766. [Google Scholar] [CrossRef] [Green Version]
- Olsson, C.; Jansson, H.; Swenson, J. The Role of Trehalose for the Stabilization of Proteins. J. Phys. Chem. B 2016, 120, 4723–4731. [Google Scholar] [CrossRef]
- Bosch, S.; De Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chrétien, D.; Jegou, D.; Bach, J.-M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef] [Green Version]
- Babazadeh, R.; Lahtvee, P.-J.; Adiels, C.B.; Goksör, M.; Nielsen, J.B.; Hohmann, S. The yeast osmostress response is carbon source dependent. Sci. Rep. 2017, 7, 990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garre, E.; Matallana, E. The three trehalases Nth1p, Nth2p and Ath1p participate in the mobilization of intracellular trehalose required for recovery from saline stress in Saccharomyces cerevisiae. Microbiology 2009, 155, 3092–3099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmud, S.A.; Nagahisa, K.; Hirasawa, T.; Yoshikawa, K.; Ashitani, K.; Shimizu, H. Effect of trehalose accumulation on response to saline stress in Saccharomyces cerevisiae. Yeast 2009, 26, 17–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwaka, S.; Kopp, M.; Holzer, H. Expression and Function of the Trehalase Genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 10193–10198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwaka, S.; Holzer, H. Molecular Biology of Trehalose and the Trehalases in the Yeast Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 1997, 58, 197–237. [Google Scholar] [CrossRef]
- Gibney, P.A.; Schieler, A.; Chen, J.C.; Rabinowitz, J.D.; Botstein, D. Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc. Natl. Acad. Sci. USA 2015, 112, 6116–6121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallerman, E.M.; Bredlau, J.P.; Camargo, L.S.A.; Dagli, M.L.Z.; Karembu, M.; Ngure, G.; Romero-Aldemita, R.; Rocha-Salavarrieta, P.J.; Tizard, M.; Walton, M.; et al. Towards progressive regulatory approaches for agricultural applications of animal biotechnology. Transgenic Res. 2022, 3, 1–33. [Google Scholar] [CrossRef]
- Wang, P.-M.; Zheng, D.-Q.; Chi, X.-Q.; Li, O.; Qian, C.-D.; Liu, T.-Z.; Zhang, X.-Y.; Du, F.-G.; Sun, P.-Y.; Qu, A.-M.; et al. Relationship of trehalose accumulation with ethanol fermentation in industrial Saccharomyces cerevisiae yeast strains. Bioresour. Technol. 2014, 152, 371–376. [Google Scholar] [CrossRef]
- Jules, M.; Beltran, G.; François, J.; Parrou, J.L. New Insights into Trehalose Metabolism by Saccharomyces cerevisiae: NTH2 Encodes a Functional Cytosolic Trehalase, and Deletion of TPS1 Reveals Ath1p-Dependent Trehalose Mobilization. Appl. Environ. Microbiol. 2008, 74, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Avila-Reyes, S.; Camacho-Diaz, B.; Acosta-Garcia, M.; Jimenez-Aparicio, A.; Hernandez-Sanchez, H. Effect of salt and sugar osmotic stress on the viability and morphology of Saccharomyces boulardii. Int. J. Environ. Agric. Biotechnol. 2016, 1, 593–602. [Google Scholar] [CrossRef]
- Logothetis, S.; Walker, G.; Nerantzis, E. Effect of salt hyperosmotic stress on yeast cell viability. Zb. Matic Srp. Prřír. Nauk. 2007, 113, 271–284. [Google Scholar] [CrossRef]
- Divate, N.R.; Chen, G.-H.; Divate, R.D.; Ou, B.-R.; Chung, Y.-C. Metabolic engineering of Saccharomyces cerevisiae for improvement in stresses tolerance. Bioengineered 2017, 8, 524–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Zhang, C.-Y.; Wu, M.-Y.; Fan, Z.-H.; Liu, S.-N.; Zhu, W.-B.; Xiao, D.-G. MAL62 overexpression and NTH1 deletion enhance the freezing tolerance and fermentation capacity of the baker’s yeast in lean dough. Microb. Cell Factories 2016, 15, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Nwaka, S.; Mechler, B.; Destruelle, M.; Holzer, H. Phenotypic features of trehalase mutants in Saccharomyces cerevisiae. FEBS Lett. 1995, 360, 286–290. [Google Scholar] [CrossRef] [Green Version]
- Mahmud, S.A.; Hirasawa, T.; Shimizu, H. Differential importance of trehalose accumulation in Saccharomyces cerevisiae in response to various environmental stresses. J. Biosci. Bioeng. 2010, 109, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Tapia, H.; Young, L.; Fox, D.; Bertozzi, C.R.; Koshland, D. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2015, 112, 6122–6127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, R.; Beach, A.; Li, K.; Haber, J. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 2017, 544, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Fernandez-Ricaud, L.; Warringer, J.; Ericson, E.; Pylvänäinen, I.; Kemp, G.J.L.; Nerman, O.; Blomberg, A. PROPHECY—a database for high-resolution phenomics. Nucleic Acids Res. 2004, 33, D369–D373. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.J.; Winters, L.; Coulson, G.E.; Clarke, K.J. Effect of Osmotic Stress on the Ultrastructure and Viability of the Yeast Saccharomyces cerevisiae. Microbiol. 1986, 132, 2023–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewald, J.C.; Kuehne, A.; Zamboni, N.; Skotheim, J.M. The Yeast Cyclin-Dependent Kinase Routes Carbon Fluxes to Fuel Cell Cycle Progression. Mol. Cell 2016, 62, 532–545. [Google Scholar] [CrossRef] [Green Version]
- Yi, C.; Wang, F.; Dong, S.; Li, H. Changes of trehalose content and expression of relative genes during the bioethanol fermentation by Saccharomyces cerevisiae. Can. J. Microbiol. 2016, 62, 827–835. [Google Scholar] [CrossRef] [Green Version]
- Zähringer, H.; Burgert, M.; Holzer, H.; Nwaka, S. Neutral trehalase Nth1p of Saccharomyces cerevisiae encoded by theNTH1gene is a multiple stress responsive protein. FEBS Lett. 1997, 412, 615–620. [Google Scholar] [CrossRef] [Green Version]
- Iwahashi, H.; Nwaka, S.; Obuchi, K. Evidence for Contribution of Neutral Trehalase in Barotolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2000, 66, 5182–5185. [Google Scholar] [CrossRef] [Green Version]
- Singer, M.A.; Lindquist, S. Thermotolerance in Saccharomyces cerevisiae: The Yin and Yang of trehalose. Trends Biotechnol. 1998, 16, 460–468. [Google Scholar] [CrossRef]
- Bravim, F.; Lippman, S.I.; da Silva, L.F.; Souza, D.T.; Fernandes, A.A.R.; Masuda, C.A.; Broach, J.R.; Fernandes, P.M.B. High hydrostatic pressure activates gene expression that leads to ethanol production enhancement in a Saccharomyces cerevisiae distillery strain. Appl. Microbiol. Biotechnol. 2013, 97, 2093–2107. [Google Scholar] [CrossRef] [Green Version]
- Botts, M.R.; Huang, M.; Borchardt, R.K.; Hull, C.M. Developmental Cell Fate and Virulence Are Linked to Trehalose Homeostasis in Cryptococcus neoformans. Eukaryot. Cell 2014, 13, 1158–1168. [Google Scholar] [CrossRef] [Green Version]
- Blevins, W.R.; Ruiz-Orera, J.; Messeguer, X.; Blasco-Moreno, B.; Villanueva-Cañas, J.L.; Espinar, L.; Díez, J.; Carey, L.B.; Albà, M.M. Uncovering de novo gene birth in yeast using deep transcriptomics. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cakiroglu, S.A.; Zaugg, J.B.; Luscombe, N.M. Backmasking in the yeast genome: Encoding overlapping information for protein-coding and RNA degradation. Nucleic Acids Res. 2016, 44, 8065–8072. [Google Scholar] [CrossRef] [Green Version]
- Orr, M.W.; Mao, Y.; Storz, G.; Qian, S.-B. Alternative ORFs and small ORFs: Shedding light on the dark proteome. Nucleic Acids Res. 2020, 48, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Beniwal, A.; Kokkiligadda, A.; Vij, S. Response and tolerance of yeast to changing environmental stress during ethanol fermentation. Process Biochem. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Pratt, P.L.; Bryce, J.H.; Stewart, G.G. The Effects of Osmotic Pressure and Ethanol on Yeast Viability and Morphology. J. Inst. Brew. 2003, 109, 218–228. [Google Scholar] [CrossRef]
- Muysson, J.; Miller, L.; Allie, R.; Inglis, D.L. The Use of CRISPR-Cas9 Genome Editing to Determine the Importance of Glycerol Uptake in Wine Yeast During Icewine Fermentation. Fermentation 2019, 5, 93. [Google Scholar] [CrossRef]
- Vilela, A. An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety. Fermentation 2021, 7, 5. [Google Scholar] [CrossRef]





| Gene | Open Reading Frames |
|---|---|
| NTH1 wild type | MSQVNTSQGPVAQGRQRRLSSLSEFNDPFSNAEVYYGPPTDPRKQKQAKPAKINRTRTMSVFDNVSPFKKTGFGKLQQTRRGSEDDTYSSSQGNRRFFIEDVDKTLNELLAAEDTDKNYQITIEDTGPKVLKVGTANSYGYKHINIRGTYMLSNLLQELTIAKSFGRHQIFLDEARINENPVNRLSRLINTQFWNSLTRRVDLNNVGEIAKDTKIDTPGAKNPRIYVPYDCPEQYEFYVQASQMHPSLKLEVEYLPKKITAEYVKSVNDTPGLLALAMEEHFNPSTGEKTLIGYPYAVPGGRFNELYGWDSYMMALGLLEANKTDVARGMVEHFIFEINHYGKILNANRSYYLCRSQPPFLTEMALVVFKKLGGRSNPDAVDLLKRAFQASIKEYKTVWTASPRLDPETGLSRYHPNGLGIPPETESDHFDTVLLPYASKHGVTLDEFKQLYNDGKIKEPKLDEFFLHDRGVRESGHDTTYRFEGVCAYLATIDLNSLLYKYEIDIADFIKEFCDDKYEDPLDHSITTSAMWKEMAKIRQEKITKYMWDDESGFFFDYNTKIKHRTSYESATTFWALWAGLATKEQAQKMVEKALPKLEMLGGLAACTERSRGPISISRPIRQWDYPFGWAPHQILAWEGLRSYGYLTVTNRLAYRWLFMMTKAFVDYNGIVVEKYDVTRGTDPHRVEAEYGNQGADFKGAATEGFGWVNASYILGLKYMNSHARRALGACIPPISFFSSLRPQERNLYGL |
| nth1 893_894insT, ORF1 | MSQVNTSQGPVAQGRQRRLSSLSEFNDPFSNAEVYYGPPTDPRKQKQAKPAKINRTRTMSVFDNVSPFKKTGFGKLQQTRRGSEDDTYSSSQGNRRFFIEDVDKTLNELLAAEDTDKNYQITIEDTGPKVLKVGTANSYGYKHINIRGTYMLSNLLQELTIAKSFGRHQIFLDEARINENPVNRLSRLINTQFWNSLTRRVDLNNVGEIAKDTKIDTPGAKNPRIYVPYDCPEQYEFYVQASQMHPSLKLEVEYLPKKITAEYVKSVNDTPGLLALAMEEHFNPSTGEKTLIGYPYAVSWW |
| nth1 893_894insT, ORF2 | MLFPGGRFNELYGWDSYMMALGLLEANKTDVARGMVEHFIFEINHYGKILNANRSYYLCRSQPPFLTEMALVVFKKLGGRSNPDAVDLLKRAFQASIKEYKTVWTASPRLDPETGLSRYHPNGLGIPPETESDHFDTVLLPYASKHGVTLDEFKQLYNDGKIKEPKLDEFFLHDRGVRESGHDTTYRFEGVCAYLATIDLNSLLYKYEIDIADFIKEFCDDKYEDPLDHSITTSAMWKEMAKIRQEKITKYMWDDESGFFFDYNTKIKHRTSYESATTFWALWAGLATKEQAQKMVEKALPKLEMLGGLAACTERSRGPISISRPIRQWDYPFGWAPHQILAWEGLRSYGYLTVTNRLAYRWLFMMTKAFVDYNGIVVEKYDVTRGTDPHRVEAEYGNQGADFKGAATEGFGWVNASYILGLKYMNSHARRALGACIPPISFFSSLRPQERNLYGL |
| NTH2 wild type | MVDFLPKVTEINPPSEGNDGEDNIKPLSSGSEQRPLKEEGQQGGRRHHRRLSSMHEYFDPFSNAEVYYGPITDPRKQSKIHRLNRTRTMSVFNKVSDFKNGMKDYTLKRRGSEDDSFLSSQGNRRFYIDNVDLALDELLASEDTDKNHQITIEDTGPKVIKVGTANSNGFKHVNVRGTYMLSNLLQELTIAKSFGRHQIFLDEARINENPVDRLSRLITTQFWTSLTRRVDLYNIAEIARDSKIDTPGAKNPRIYVPYNCPEQYEFYIQASQMNPSLKLEVEYLPKDITAEYVKSLNDTPGLLALAMEEHVNPSTGERSLVGYPYAVPGGRFNELYGWDSYLMALGLIESNKVDVARGMVEHFIFEIDHYSKILNANRSYYLCRSQPPFLTDMALLVFEKIGGKNNPNAIQLLKRAFRAAIKEYKEVWMSSPRLDSLTGLSCYHSDGIGIPPETEPDHFDTILLPYAEKYNVTLEKLRYLYNEGMIKEPKLDAFFLHDRAVRESGHDTTYRFEGVCAYLATIDLNSLLYKYEKDIAFVIKEYFGNEYKDENDGTVTDSEHWEELAELRKTRINKYMWDEDSGFFFYYNTKLKCRTSYESATTFWSLWAGLATEEQAKITVEKALPQLEMLGGLVACTEKSRGPISIDRPIRQWDYPFGWAPHQILAWKGLSAYGYQQVATRLAYRWLYMITKSFVDYNGMVVEKYDVTRGTDPHRVDAEYGNQGADFKGVATEGFGWVNTSYLLGLKYMNNHARRALAACSPPLPFFNSLKPSEKKLYYL |
| nth2 1271_1272delTA, ORF1 | MVDFLPKVTEINPPSEGNDGEDNIKPLSSGSEQRPLKEEGQQGGRRHHRRLSSMHEYFDPFSNAEVYYGPITDPRKQSKIHRLNRTRTMSVFNKVSDFKNGMKDYTLKRRGSEDDSFLSSQGNRRFYIDNVDLALDELLASEDTDKNHQITIEDTGPKVIKVGTANSNGFKHVNVRGTYMLSNLLQELTIAKSFGRHQIFLDEARINENPVDRLSRLITTQFWTSLTRRVDLYNIAEIARDSKIDTPGAKNPRIYVPYNCPEQYEFYIQASQMNPSLKLEVEYLPKDITAEYVKSLNDTPGLLALAMEEHVNPSTGERSLVGYPYAVPGGRFNELYGWDSYLMALGLIESNKVDVARGMVEHFIFEIDHYSKILNANRSYYLCRSQPPFLTDMALLVFEKIGGKNNPNAIQLLKRAFRAAIKE |
| nth2 1271_1272delTA, ORF2 | MSSPRLDSLTGLSCYHSDGIGIPPETEPDHFDTILLPYAEKYNVTLEKLRYLYNEGMIKEPKLDAFFLHDRAVRESGHDTTYRFEGVCAYLATIDLNSLLYKYEKDIAFVIKEYFGNEYKDENDGTVTDSEHWEELAELRKTRINKYMWDEDSGFFFYYNTKLKCRTSYESATTFWSLWAGLATEEQAKITVEKALPQLEMLGGLVACTEKSRGPISIDRPIRQWDYPFGWAPHQILAWKGLSAYGYQQVATRLAYRWLYMITKSFVDYNGMVVEKYDVTRGTDPHRVDAEYGNQGADFKGVATEGFGWVNTSYLLGLKYMNNHARRALAACSPPLPFFNSLKPSEKKLYYL |
| Strain 1 | Intracellular Content of Trehalose | |
|---|---|---|
| 0 M NaCl | 0.85 M NaCl | |
| S. cerevisiae CENPK2 (control) | (150 ± 22) mg 100 mL−1 | (118 ± 18) mg/100 mL |
| S. cerevisiae CENPK2 nth2 1271_1272delTA | (139 ± 21) mg 100 mL−1 | (107 ± 16) mg/100 mL |
| S. cerevisiae CENPK2 nth1 893_894insT | (34.8 ± 5.2) mg 100 mL−1 | (33.8 ± 5.1) mg/100 mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Soto, A.; Delgado-Navarro, J.P.; Benavides-Acevedo, M.; Paniagua, S.A.; Gatica-Arias, A. NTH2 1271_1272delTA Gene Disruption Results in Salt Tolerance in Saccharomyces cerevisiae. Fermentation 2022, 8, 166. https://doi.org/10.3390/fermentation8040166
Hernández-Soto A, Delgado-Navarro JP, Benavides-Acevedo M, Paniagua SA, Gatica-Arias A. NTH2 1271_1272delTA Gene Disruption Results in Salt Tolerance in Saccharomyces cerevisiae. Fermentation. 2022; 8(4):166. https://doi.org/10.3390/fermentation8040166
Chicago/Turabian StyleHernández-Soto, Alejandro, José Pablo Delgado-Navarro, Miguel Benavides-Acevedo, Sergio A. Paniagua, and Andres Gatica-Arias. 2022. "NTH2 1271_1272delTA Gene Disruption Results in Salt Tolerance in Saccharomyces cerevisiae" Fermentation 8, no. 4: 166. https://doi.org/10.3390/fermentation8040166