Unraveling How Candida albicans Forms Sexual Biofilms
Abstract
:1. Introduction
2. The White–Opaque Transition
3. Pheromone Signaling and Response
3.1. Mating Pheromones
3.2. Pheromone-Signaling Pathway Control
3.3. Differences Between the White and Opaque Cell Pheromone Responses
4. Conventional and Sexual Biofilms
4.1. Properties of Conventional and Sexual Biofilms Compared
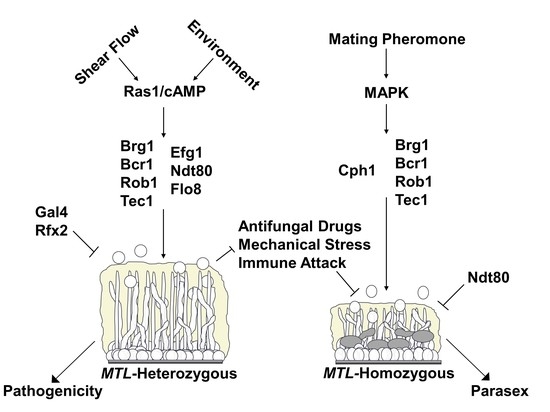
4.2. Genetic Regulation of Conventional and Sexual Biofilms
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kolter, R.; Greenberg, E.P. Microbial sciences: The superficial life of microbes. Nature 2006, 441, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single-and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2018, 16, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, R.P. Nosocomial Candidemia: Risk Factors and Attributable Mortality. Clin. Infect. Dis. 1995, 20, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.; Walsh, T.J.; Sobel, J.D.; Filler, S.G.; Pappas, P.G.; Dismukes, W.E.; Edwards, J.E. Practice Guidelines for the treatment of candidiasis. Clin. Infect. Dis. 2000, 30, 662–678. [Google Scholar] [CrossRef]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Kullber, B.J.; Oude Lashof, A.M. Epidemiology of opportunisitic invasive mycoses. Eur. J. Med. Res. 2002, 7, 183–191. [Google Scholar]
- Weig, M.; Gross, U.; Mühlschlegel, F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998, 6, 468–470. [Google Scholar] [CrossRef]
- Daniels, K.J.; Srikantha, T.; Lockhart, S.R.; Pujol, C.; Soll, D.R. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006, 25, 2240–2252. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.N.; Daniels, K.J.; Pujol, C.; Srikantha, T.; Soll, D.R. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot. Cell 2013, 12, 1120–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganguly, S.; Mitchell, A.P. Mucosal biofilms of Candida albicans. Curr. Opin. Microbiol. 2011, 14, 380–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumamoto, C.A. Candida biofilms. Curr. Opin. Microbiol. 2002, 5, 608–611. [Google Scholar] [CrossRef]
- Kumamoto, C.A. Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 2011, 14, 386–391. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, M.J.; Volz, P.A. Ecology of Candida albicans gut colonization: Inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 1985, 49, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Köhler, J.R.; Casadevall, A.; Perfect, J. The Spectrum of Fungi That Infects Humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019273. [Google Scholar] [CrossRef] [Green Version]
- Lohse, M.B.; Johnson, A.D. White-opaque switching in Candida albicans. Curr. Opin. Microbiol. 2009, 12, 650–654. [Google Scholar] [CrossRef] [Green Version]
- Slutsky, B.; Staebell, M.; Anderson, J.; Risen, L.; Pfaller, M.; Soll, D.R. “White-opaque transition”: A second high-frequency switching system in Candida albicans. J. Bacteriol. 1987, 169, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Yi, S.; Sahni, N.; Daniels, K.J.; Srikantha, T.; Soll, D.R. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010, 6, e1000806. [Google Scholar] [CrossRef]
- Xie, J.; Tao, L.; Nobile, C.J.; Tong, Y.; Guan, G.; Sun, Y.; Cao, C.; Hernday, A.D.; Johnson, A.D.; Zhang, L.; et al. White-Opaque Switching in Natural MTLa/α Isolates of Candida albicans: Evolutionary Implications for Roles in Host Adaptation, Pathogenesis, and Sex. PLoS Biol. 2013, 11, e1001525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuch, B.B.; Mitrovich, Q.M.; Homann, O.R.; Hernday, A.D.; Monighetti, C.K.; de La Vega, F.M.; Johnson, A.D. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010, 6, e1001070. [Google Scholar] [CrossRef] [Green Version]
- Lan, C.Y.; Newport, G.; Murillo, L.A.; Jones, T.; Scherer, S.; Davis, R.W.; Agabian, N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 2002, 99, 14907–14912. [Google Scholar] [CrossRef] [Green Version]
- Tsong, A.E.; Miller, M.G.; Raisner, R.M.; Johnson, A.D. Evolution of a Combinatorial Transcriptional Circuit: A Case Study in Yeasts. Cell 2003, 115, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Ene, I.V.; Lohse, M.B.; Vladu, A.V.; Morschhäuser, J.; Johnson, A.D.; Bennett, R.J. Phenotypic profiling reveals that Candida albicans opaque cells represent a metabolically specialized cell state compared to default white cells. MBio 2016, 7, e01269-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, H.; Hernday, A.D.; Hirakawa, M.P.; Johnson, A.D.; Bennett, R.J. Candida albicans White and Opaque Cells Undergo Distinct Programs of Filamentous Growth. PLoS Pathog. 2013, 9, e1003210. [Google Scholar] [CrossRef] [Green Version]
- Dumitru, R.; Navarathna, D.H.M.L.P.; Semighini, C.P.; Elowsky, C.G.; Dumitru, R.V.; Dignard, D.; Whiteway, M.; Atkin, A.L.; Nickerson, K.W. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot. Cell 2007, 6, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 2012, 3, 251–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez-Zavala, B.; Reuß, O.; Park, Y.; Ohlsen, K.; Morschhäuser, J. Environmental Induction of White–Opaque Switching in Candida albicans. PLoS Pathog. 2008, 4, e1000089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Srikantha, T.; Sahni, N.; Yi, S.; Soll, D.R. Report CO(2) Regulates White-to-Opaque Switching in Candida albicans. Curr. Biol. 2009, 19, 330–334. [Google Scholar] [CrossRef] [Green Version]
- Alby, K.; Bennett, R.J. Stress-Induced Phenotypic Switching in Candida albicans. Mol. Biol. Cell 2009, 20, 3178–3191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, J.; Wessels, D.; Lockhart, S.R.; Soll, D.R. Release of a Potent Polymorphonuclear Leukocyte Chemoattractant Is Regulated by White-Opaque Switching in Candida albicans. Infect. Immun. 2004, 72, 667–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.G.; Johnson, A.D. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 2002, 110, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, S.R.; Pujol, C.; Daniels, K.J.; Miller, M.G.; Johnson, A.D.; Pfaller, M.A.; Soll, D.R. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 2002, 162, 737–745. [Google Scholar]
- Hull, C.M.; Johnson, A.D. Identification of a Mating Type–Like Locus in the Asexual Pathogenic Yeast Candida albicans. Science 1999, 285, 1271–1275. [Google Scholar] [CrossRef]
- Huang, G.; Wang, H.; Chou, S.; Nie, X.; Chen, J.; Liu, H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. USA 2006, 103, 12813–12818. [Google Scholar] [CrossRef] [Green Version]
- Hernday, A.D.; Lohse, M.B.; Fordyce, P.M.; Nobile, C.J.; DeRisi, J.L.; Johnson, A.D. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol. 2013, 90, 22–35. [Google Scholar]
- Pendrak, M.L.; Yan, S.S.; Roberts, D.D. Hemoglobin regulates expression of an activator of mating-type locus α genes in Candida albicans. Eukaryot. Cell 2004, 3, 764–775. [Google Scholar] [CrossRef] [Green Version]
- Pendrak, M.L.; Yan, S.S.; Roberts, D.D. Sensing the host environment: Recognition of hemoglobin by the pathogenic yeast Candida albicans. Arch. Biochem. Biophys. 2004, 426, 148–156. [Google Scholar] [CrossRef]
- Sun, Y.; Gadoury, C.; Hirakawa, M.P.; Bennett, R.J.; Harcus, D.; Marcil, A. Deletion of a Yci1 Domain Protein of Candida albicans Allows Homothallic Mating in MTL Heterozygous Cells. MBio 2016, 7, e00465-16. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, M.P.; Martinez, D.A.; Sakthikumar, S.; Anderson, M.Z.; Berlin, A.; Gujja, S.; Zeng, Q.; Zisson, E.; Wang, J.M.; Greenberg, J.M.; et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015, 25, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magee, B.B.; Magee, P.T. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 2000, 289, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2011, 2, e00129-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, T.Y.; Chang, F.M.; Cheng, W.N.; Lara, A.; Chou, M.L.; Lee, W.F.; Lee, K.C.; Lin, C.T.; Lee, W.S.; Yu, F.L.; et al. Fluconazole induces rapid high-frequency MTL homozygosis with microbiological polymorphism in Candida albicans. J. Microbiol. Immunol. Infect. 2017, 50, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Hilton, C.; Markie, D.; Corner, B.; Rikkerink, E.; Poulter, R. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 1985, 200, 162. [Google Scholar] [CrossRef]
- Berman, J.; Hadany, L. Does stress induce (para) sex? Implications for Candida albicans evolution. Trends Genet. 2012, 28, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Hull, C.M. Evidence for Mating of the “Asexual” Yeast Candida albicans in a Mammalian Host. Science 2000, 289, 307–310. [Google Scholar] [CrossRef]
- Bennett, R.J.; Johnson, A.D. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003, 22, 2505–2515. [Google Scholar] [CrossRef] [Green Version]
- Bennett, R.J. The parasexual lifestyle of Candida albicans. Curr. Opin. Microbiol. 2015, 28, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Alby, K.; Schaefer, D.; Bennett, R.J. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 2009, 460, 890–893. [Google Scholar] [CrossRef] [Green Version]
- Soll, D.R.; Lockhart, S.R.; Zhao, R. Relationship between switching and mating in Candida albicans. Eukaryot. Cell 2003, 2, 390–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockhart, S.R.; Zhao, R.; Daniels, K.J.; Soll, D.R. α-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2003, 2, 847–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockhart, S.R.; Daniels, K.J.; Zhao, R.; Wessels, D.; Soll, D.R. Cell Biology of Mating in Candida albicans. Eukaryot. Cell 2003, 2, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Forche, A.; Alby, K.; Schaefer, D.; Johnson, A.D.; Berman, J.; Bennett, R.J. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008, 6, e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, M.A.; Paulson, C.; Dudley, A.; Berman, J. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics 2015, 200, 781–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, G.; Tao, L.; Yue, H.; Liang, W.; Gong, J.; Bing, J.; Zheng, Q.; Veri, A.O.; Fan, S.; Robbins, N.; et al. Environment-induced same-sex mating in the yeast Candida albicans through the Hsf1–Hsp90 pathway. PLoS Biol. 2019, 17, e2006966. [Google Scholar] [CrossRef] [Green Version]
- Sahni, N.; Yi, S.; Pujol, C.; Soll, D.R. The white cell response to pheromone is a general characteristic of Candida albicans strains. Eukaryot. Cell 2009, 8, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.H.; Kabrawala, S.; Fox, E.P.; Nobile, C.J.; Johnson, A.D.; Bennett, R.J. Genetic Control of Conventional and Pheromone-Stimulated Biofilm Formation in Candida albicans. PLoS Pathog. 2013, 9, e1003305. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.; Sahni, N.; Daniels, K.J.; Lu, K.L.; Srikantha, T.; Huang, G.; Garnaas, A.M.; Soll, D.R. Alternative mating type configurations (a/α versus a/a or α/α) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol. 2011, 9, e1001117. [Google Scholar] [CrossRef] [Green Version]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Dignard, D.; El-Naggar, A.L.; Logue, M.E.; Butler, G.; Whiteway, M. Identification and characterization of MFAl, the gene encoding Candida albicans a-factor pheromone. Eukaryot. Cell 2007, 6, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, R.; Uhl, M.A.; Miller, M.G.; Johnson, A. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 2003, 23, 8189–8201. [Google Scholar] [CrossRef] [Green Version]
- Panwar, S.L.; Legrand, M.; Dignard, D.; Whiteway, M.; Magee, P.T. MFα1, the Gene Encoding the α Mating Pheromone of Candida albicans. Eukaryot. Cell. 2003, 2, 1350–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julius, D.; Brake, A.; Blair, L.; Kunisawa, R.; Thorner, J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-α-factor. Cell 1984, 37, 1075–1089. [Google Scholar] [CrossRef]
- Newport, G.; Agabian, N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol. Chem. 1997, 272, 28954–28961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julius, D.; Blair, L.; Brake, A.; Sprague, G.; Thorner, J. Yeast α factor is processed from a larger precursor polypeptide: The essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 1983, 32, 839–852. [Google Scholar] [CrossRef]
- Bautista-Muñoz, C.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Analysis and expression of STE13ca gene encoding a putative X-prolyl dipeptidyl aminopeptidase from Candida albicans. FEMS Immunol. Med. Microbiol. 2005, 45, 459–469. [Google Scholar] [CrossRef]
- Bennett, R.J.; Johnson, A.D. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of Candida albicans. Mol. Microbiol. 2006, 62, 100–119. [Google Scholar] [CrossRef]
- Chen, P.; Sapperstein, S.K.; Choi, J.D.; Michaelis, S. Biogenesis of the Saccharomyces cerevisiae Mating Pheromone a-Factor. J. Cell Biol. 1997, 136, 251–269. [Google Scholar] [CrossRef] [Green Version]
- Magee, B.B.; Legrand, M.; Alarco, A.M.; Raymond, M.; Magee, P.T. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 2002, 46, 1345–1351. [Google Scholar] [CrossRef]
- Raymond, M.; Dignard, D.; Alarco, A.M.; Mainville, N.; Magee, B.B.; Thomas, D.Y. A Ste6p/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 27, 587–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, D.; Côte, P.; Whiteway, M.; Bennett, R.J. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot. Cell 2007, 6, 907–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Chen, J.; Lane, S.; Liu, H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 2002, 46, 1335–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, S.; Sahni, N.; Daniels, K.J.; Pujol, C.; Srikantha, T.; Soll, D.R. The Same Receptor, G Protein, and Mitogen-activated Protein Kinase Pathway Activate Different Downstream Regulators in the Alternative White and Opaque Pheromone Responses of Candida albicans. Mol. Biol. Cell 2008, 19, 957–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.H.; Choi, A.; Bennett, R.J. Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Mol. Biol. Cell 2011, 22, 4918–4930. [Google Scholar] [CrossRef] [PubMed]
- Dignard, D.; André, D.; Whiteway, M. Heterotrimeric G-protein subunit function in Candida albicans: Both the α and β subunits of the pheromone response G protein are required for mating. Eukaryot. Cell 2008, 7, 1591–1599. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.; Sahni, N.; Daniels, K.J.; Lu, K.L.; Huang, G.; Garnaas, A.M.; Pujol, C.; Srikantha, T.; Soll, D.R. Utilization of the Mating Scaffold Protein in the Evolution of a New Signal Transduction Pathway for Biofilm Development. MBio 2011, 2, e00237-10. [Google Scholar] [CrossRef] [Green Version]
- Rastghalam, G.; Omran, R.P.; Alizadeh, M.; Fulton, D.; Mallick, J.; Whiteway, M. MAP Kinase Regulation of the Candida albicans Pheromone Pathway. mSphere 2019, 4, e00598-18. [Google Scholar] [CrossRef] [Green Version]
- Scaduto, C.M.; Kabrawala, S.; Thomson, G.J.; Scheving, W.; Ly, A.; Anderson, M.Z.; Whiteway, M.; Bennett, R.J. Epigenetic control of pheromone MAPK signaling determines sexual fecundity in Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Dohlman, H.G.; Song, J.; Apanovitch, D.M.; DiBello, P.R.; Gillen, K.M. Regulation of G protein signalling in yeast. Semin. Cell Dev. Biol. 1998, 9, 135–141. [Google Scholar] [CrossRef]
- Ramírez-Zavala, B.; Weyler, M.; Gildor, T.; Schmauch, C.; Kornitzer, D.; Arkowitz, R.; Morschhäuser, J. Activation of the Cph1-Dependent MAP Kinase Signaling Pathway Induces White-Opaque Switching in Candida albicans. PLoS Pathog. 2013, 9, e1003696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, R.L.; Fink, G.R. Elements of a single map kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994, 8, 2974–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herskowitz, I. MAP Kinase Pathways in Yeast: For Mating and More. Cell 1995, 80, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Hartwell, L.H. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp. Cell Res. 1973, 76, 111–117. [Google Scholar] [CrossRef]
- Nantel, A.; Dignard, D.; Bachewich, C.; Harcus, D.; Marcil, A.; Bouin, A.P.; Sensen, C.W.; Hogues, H.; van het Hoog, M.; Gordon, P.; et al. Transcription Profiling of Candida albicans Cells Undergoing Yeast-to-Hyphal Transistion. Mol. Biol. Cell 2002, 13, 3452–3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, K.J.; Park, Y.N.; Srikantha, T.; Pujol, C.; Soll, D.R. Impact of Environmental Conditions on the Form and Function of Candida albicans Biofilms. Eukaryot. Cell 2013, 12, 1389–1402. [Google Scholar] [CrossRef] [Green Version]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramanian, A.; Köehler, J.; Kadosh, D.; Lopez-ribot, J.L. Dispersion as an Important Step in the Candida albicans Biofilm Developmental Cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of Fluconazole Resistance in Candida albicans Biofilms: Phase-Specific Role of Efflux Pumps and Membrane Sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [Green Version]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [Green Version]
- Tao, L.; Cao, C.; Liang, W.; Guan, G.; Zhang, Q.; Nobile, C.J.; Huang, G. White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in Candida albicans. PLoS Genet. 2014, 10, e1004737. [Google Scholar] [CrossRef] [Green Version]
- Lohse, M.B.; Johnson, A.D. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE 2008, 3, e0001473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kean, R.; Delaney, C.; Rajendran, R.; Sherry, L.; Metcalfe, R.; Thomas, R.; Mclean, W.; Williams, C.; Ramage, G. Gaining Insights from Candida Biofilm Heterogeneity: One Size Does Not Fit All. J. Fungi 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [Green Version]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.J.; Alsad, L.; Vogel, F.; Koppar, S.; Nevarez, L.; Auguste, F.; Seymour, J.; Syed, A.; Christoph, K.; Loomis, J.S. Interactions between Candida albicans and Staphylococcus aureus within mixed species biofilms. Bios 2013, 84, 30–39. [Google Scholar] [CrossRef]
- Peters, B.M.; Ovchinnikova, E.S.; Krom, B.P.; Schlecht, L.M.; Zhou, H.; Hoyer, L.L.; Busscher, H.J.; van der Mei, H.C.; Jabra-Rizk, M.A.; Shirtliff, M.E. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 2012, 158, 2975–2986. [Google Scholar] [CrossRef] [Green Version]
- Peters, B.M.; Jabra-Rizk, M.A.; Scheper, M.A.; Leid, J.G.; Costerton, J.W.; Shirtliff, M.E. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Andes, D.; van Dijck, P.; Jabra-Rizk, M.A. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio 2016, 7, e01365-16. [Google Scholar] [CrossRef] [Green Version]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, A.K.; Hogan, D.A. Candida albicans: Molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol. Rev. 2014, 28, 85–96. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Wickes, B.L.; López-ribot, J.L. Inhibition of Candida albicans Biofilm Formation by Farnesol, a Quorum-Sensing Molecule. Appl. Environ. Microbiol. 2002, 68, 5459–5463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, D.K.; Grahl, N.; Okegbe, C.; Dietrich, L.E.P.; Jacobs, N.J.; Hogan, A. Control of Candida albicans Metabolism and Biofilm Formation by Pseudomonas aeruginosa Phenazines. MBio 2013, 4, e00526-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kernien, J.F.; Snarr, B.D.; Sheppard, D.C.; Nett, J.E. The interface between Fungal Biofilms and Innate Immunity. Front. Immunol. 2018, 8, 1968. [Google Scholar] [CrossRef] [Green Version]
- Brown, G.D. Innate Antifungal Immunity: The Key Role of Phagocytes. Annu. Rev. Immunol. 2011, 29, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans and hyphal forms. Cell. Microbiol. 2006, 8, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Cabezas-Olcoz, J.; Kernien, J.F.; Wang, S.X.; Beebe, D.J.; Huttenlocher, A.; Ansari, H.; Nett, J.E. The Extracellular Matrix of Candida albicans Biofilms Impairs Formation of Neutrophil Extracellular Traps. PLoS Pathog. 2016, 12, e1005884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J. Infect. Dis. 2012, 206, 1936–1945. [Google Scholar] [CrossRef] [Green Version]
- Chandra, J.; McCormick, T.S.; Imamura, Y.; Mukherjee, P.K.; Ghannoum, M.A. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect. Immun. 2007, 75, 2612–2620. [Google Scholar] [CrossRef] [Green Version]
- Rocha, F.A.C.; Alves, A.M.C.V.; Rocha, M.F.G.; Cordeiro, R.D.A.; Brilhante, R.S.N.; Pinto, A.C.M.D.; Nunes, R.D.M.; Girão, V.C.C.; Sidrim, J.J.C. Tumor necrosis factor prevents Candida albicans biofilm formation. Sci. Rep. 2017, 7, 1206. [Google Scholar] [CrossRef] [Green Version]
- Gantner, B.N.; Simmons, R.M.; Underhill, D.M. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO 2005, 24, 1277–1286. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Wagner, A.S.; Tams, R.N.; Eyer, J.E.; Kauffman, S.J.; Gann, E.R.; Fernandez, E.J.; Reynolds, T.B. Lrg1 Regulates β (1,3)-Glucan Masking in Candida albicans through the Cek1 MAP Kinase Pathway. MBio 2019, 10, e01767-19. [Google Scholar] [CrossRef] [Green Version]
- Inglis, D.O.; Sherlock, G. Ras signaling gets fine-tuned: Regulation of multiple pathogenic traits of Candida albicans. Eukaryot. Cell 2013, 12, 1316–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Huang, Q.; Wei, Y.; Wang, Y.; Du, H. Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol. Microbiol. 2019, 111, 6–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, T.R.; Johnson, A.D. Making sense of transcription networks. Cell 2015, 161, 714–723. [Google Scholar] [CrossRef] [Green Version]
- Nobile, C.J.; Nett, J.E.; Hernday, A.D.; Homann, O.R.; Deneault, J.; Nantel, A.; Andes, D.R.; Johnson, A.D.; Mitchell, A.P. Biofilm Matrix Regulation by Candida albicans Zap1. PLoS Biol. 2009, 7, e1000133. [Google Scholar] [CrossRef] [Green Version]
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Ross, K.M.; Andes, D.R. Interface of Candida albicans Biofilm Matrix-Associated Drug Resistance and Cell Wall Integrity Regulation. Eukaryot. Cell 2011, 10, 1660–1669. [Google Scholar] [CrossRef] [Green Version]
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Andes, D.R. Genetic Basis of Candida Biofilm Resistance Due to Drug-Sequestering Matrix Glucan. J. Infect. Dis. 2010, 202, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Daniels, K.J.; Srikantha, T.; Pujol, C.; Park, Y.N.; Soll, D.R. Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot. Cell 2015, 14, 228–240. [Google Scholar] [CrossRef] [Green Version]
- Sahni, N.; Yi, S.; Daniels, K.J.; Huang, G.; Srikantha, T.; Soll, D.R. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: Insights into the evolution of new signal transduction pathways. PLoS Biol. 2010, 8, e1000363. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perry, A.M.; Hernday, A.D.; Nobile, C.J. Unraveling How Candida albicans Forms Sexual Biofilms. J. Fungi 2020, 6, 14. https://doi.org/10.3390/jof6010014
Perry AM, Hernday AD, Nobile CJ. Unraveling How Candida albicans Forms Sexual Biofilms. Journal of Fungi. 2020; 6(1):14. https://doi.org/10.3390/jof6010014
Chicago/Turabian StylePerry, Austin M., Aaron D. Hernday, and Clarissa J. Nobile. 2020. "Unraveling How Candida albicans Forms Sexual Biofilms" Journal of Fungi 6, no. 1: 14. https://doi.org/10.3390/jof6010014