Forecasting Zoonotic Infectious Disease Response to Climate Change: Mosquito Vectors and a Changing Environment
Abstract
:1. Introduction
2. Changing Mosquito Vector Biology and Range Expansion
3. Epidemiological Modeling of Vector-Borne Infectious Diseases
4. Earth System Modeling: The State of Climate Forecasting
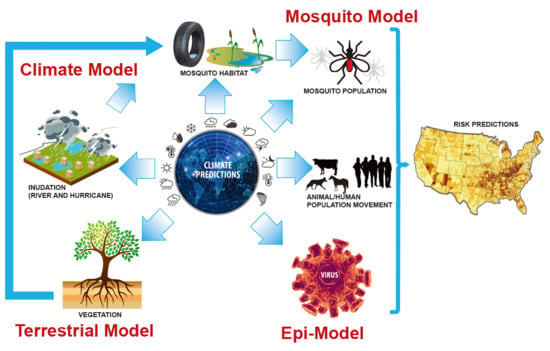
5. Bridging the Gap Between Epidemiological and Earth System Modeling
6. Data Fusion and Data Requirements
7. Assumptions or Challenges of Disease Forecasting and Modeling in Animals and Humans
8. Concluding Remarks: What is Needed Now and in the Future for Forecasting the Impacts of Climate Change on Infectious Diseases
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. A Global Brief on Vector-Borne Diseases; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I.; Bedi, N.; Bensenor, I.M.; Castañeda-Orjuela, C.A.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet. Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef]
- Vasilakis, N.; Cardosa, J.; Hanley, K.A.; Holmes, E.C.; Weaver, S.C. Fever from the forest: Prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 2011, 9, 532–541. [Google Scholar] [CrossRef]
- Wikel, S. Ticks and Tick-Borne Infections: Complex Ecology, Agents, and Host Interactions. Vet. Sci. 2018, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Alto, B.W.; Juliano, S.A. Precipitation and temperature effects on populations of Aedes albopictus (Diptera: Culicidae): Implications for range expansion. J. Med. Entomol. 2001, 38, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Afrane, Y.A.; Githeko, A.K.; Yan, G. The ecology of Anopheles mosquitoes under climate change: Case studies from the effects of deforestation in East African highlands. Ann. N. Y. Acad. Sci. 2012, 1249, 204–210. [Google Scholar] [CrossRef]
- Johnson, B.J.; Sukhdeo, M.V.K. Drought-induced amplification of local and regional West Nile virus infection rates in New Jersey. J. Med. Entomol. 2013, 50, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.M.; Obermayr, U.; Fischer, D.; Kreyling, J.; Beierkuhnlein, C. Low-temperature threshold for egg survival of a post-diapause and non-diapause European aedine strain, Aedes albopictus (Diptera: Culicidae). Parasit. Vectors 2012, 5, 100. [Google Scholar] [CrossRef]
- Rochlin, I.; Ninivaggi, D.V.; Hutchinson, M.L.; Farajollahi, A. Climate change and range expansion of the Asian tiger mosquito (Aedes albopictus) in Northeastern USA: Implications for public health practitioners. PLoS ONE 2013, 8, e60874. [Google Scholar] [CrossRef]
- Liu, H.; Li, M.-H.; Zhai, Y.-G.; Meng, W.-S.; Sun, X.-H.; Cao, Y.-X.; Fu, S.-H.; Wang, H.-Y.; Xu, L.-H.; Tang, Q.; et al. Banna Virus, China, 1987–2007. Emerg. Infect. Dis. 2010, 16, 514–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okeoma, C.M. Chikungunya Virus; Okeoma, C.M., Ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-42956-4. [Google Scholar]
- Sudeep, A.B.; Parashar, D. Chikungunya: An overview. J. Biosci. 2008, 33, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-J.; Putnak, R. Dengue and the Dengue Viruses. Perspec. Med. Virol. 2006, 16, 269–298. [Google Scholar]
- Barr, K.L.; Anderson, B.D.; Heil, G.L.; Friary, J.A.; Gray, G.C.; Focks, D.A. Focks Dengue serotypes 1–4 exhibit unique host specificity in vitro. Virus Adapt. Treat. 2012, 4, 65. [Google Scholar] [CrossRef]
- Sukhralia, S.; Verma, M.; Gopirajan, S.; Dhanaraj, P.S.; Lal, R.; Mehla, N.; Kant, C.R. From dengue to Zika: The wide spread of mosquito-borne arboviruses. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Kartashev, V.; Afonin, A.; González-Miguel, J.; Sepúlveda, R.; Simón, L.; Morchón, R.; Simón, F. Regional Warming and Emerging Vector-Borne Zoonotic Dirofilariosis in the Russian Federation, Ukraine, and Other Post-Soviet States from 1981 to 2011 and Projection by 2030. Biomed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, P.K.; Rymkiewicz, G.; Golebiewska, J.; Nestoros, N.; Romejko-Jarosinska, J.; Zarnowska-Prymek, H.; Masny, A.; Palucki, J.; Cielecka, D. The first case of human autochtonous subconjunctival dirofilariosis in Poland and MALT lymphoma as possible consequence of this parasitosis. Infect. Agent Cancer 2015, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Estep, L.K.; McClure, C.J.W.; Vander Kelen, P.; Burkett-Cadena, N.D.; Sickerman, S.; Hernandez, J.; Jinright, J.; Hunt, B.; Lusk, J.; Hoover, V.; et al. Risk of exposure to eastern equine encephalomyelitis virus increases with the density of northern cardinals. PLoS ONE 2013, 8, e57879. [Google Scholar] [CrossRef] [PubMed]
- Silverman, M.A.; Misasi, J.; Smole, S.; Feldman, H.A.; Cohen, A.B.; Santagata, S.; Mcmanus, M.; Ahmed, A.A. Eastern equine encephalitis in children, Massachusetts and New Hampshire, USA, 1970-2010. Emerg. Infect. Dis. 2013, 19, 194–202. [Google Scholar] [CrossRef] [PubMed]
- OIE World Organization for Animal Health. Technical Disease Cards—Vesicular Stomatitis. 2017. Available online: http://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/VESICULAR_STOMATITIS.pdf (accessed on 15 December 2018).
- Parmet, S.; Lynm, C.; RM, G. Malaria. JAMA 2010, 304, 2084. [Google Scholar] [CrossRef]
- Pergantas, P.; Tsatsaris, A.; Malesios, C.; Kriparakou, G.; Demiris, N.; Tselentis, Y. A spatial predictive model for malaria resurgence in central Greece integrating entomological, environmental and social data. PLoS ONE 2017, 12, e0178836. [Google Scholar] [CrossRef]
- Hassing, R.-J.; Leparc-Goffart, I.; Blank, S.N.; Thevarayan, S.; Tolou, H.; van Doornum, G.; van Genderen, P.J. Imported Mayaro virus infection in the Netherlands. J. Infect. 2010, 61, 343–345. [Google Scholar] [CrossRef]
- Niven, D.J.; Afra, K.; Iftinca, M.; Tellier, R.; Fonseca, K.; Kramer, A.; Safronetz, D.; Holloway, K.; Drebot, M.; Johnson, A.S. Fatal infection with murray valley encephalitis virus imported from Australia to Canada, 2011. Emerg. Infect. Dis. 2017, 23, 280–283. [Google Scholar] [CrossRef]
- Brault, A.C.; Tesh, R.B.; Powers, A.M.; Weaver, S.C. Re-emergence of chikungunya and o’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar]
- Travassos Da Rosa, J.F.; De Souza, W.M.; De Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Teixeira Nunes, M.R. Oropouche virus: Clinical, epidemiological, and molecular aspects of a neglected orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [PubMed]
- Henry, R.; Murphy, F.A. Etymologia: Oropouche Virus. Emerg. Infect. Dis. 2018, 24, 30329. [Google Scholar] [CrossRef]
- Berger, S. Rift Valley Fever: Global Status; Gideon Informatics: Los Angeles, CA, USA, 2014. [Google Scholar]
- Kading, R.C.; Kityo, R.M.; Mossel, E.C.; Borland, E.M.; Nakayiki, T.; Nalikka, B.; Nyakarahuka, L.; Ledermann, J.P.; Panella, N.A.; Gilbert, A.T.; et al. Neutralizing antibodies against flaviviruses, Babanki virus, and Rift Valley fever virus in Ugandan bats. Infect. Ecol. Epidemiol. 2018, 8, 1439215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottendorfer, C.L.; Ambrose, J.H.; White, G.S.; Unnasch, T.R.; Stark, L.M. Isolation of Genotype V St. Louis Encephalitis Virus in Florida. Emerg. Infect. Dis. 2009, 15, 604–606. [Google Scholar] [CrossRef] [Green Version]
- Diaz, A.; Coffey, L.L.; Burkett-Cadena, N.; Day, J.F. Reemergence of St. Louis Encephalitis Virus in the Americas. Emerg. Infect. Dis. 2018, 24, 2150. [Google Scholar] [CrossRef]
- White, S.K.; Lednicky, J.A.; Okech, B.A.; Morris, J.G.; Dunford, J.C. Spondweni virus in field-caught culex quinquefasciatus mosquitoes, Haiti, 2016. Emerg. Infect. Dis. 2018, 24, 1765–1767. [Google Scholar] [CrossRef]
- OIE World Organization for Animal Health. Technical Disease Cards—Trypanosomosis (Tsetse-Transmitted). 2013. Available online: www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/TRYPANO_TSETSE.pdf (accessed on 15 December 2018).
- Vazquez, A.; Jimenez-Clavero, M.; Franco, L.; Donoso-Mantke, O.; Sambri, V.; Niedrig, M.; Zeller, H.; Tenorio, A. Usutu virus: Potential risk of human disease in Europe. Euro Surveill. 2011, 16, 19935. [Google Scholar]
- Cadar, D.; Becker, N.; de Mendonca Campos, R.; Börstler, J.; Jöst, H.; Schmidt-Chanasit, J. Usutu Virus in Bats. Emerg. Infect. Dis. 2014, 20, 2013–2015. [Google Scholar]
- Reiss, C.S. Neurotropic Viral Infections; Cambridge University Press: Cambridge, UK, 2008; ISBN 9780511541728. [Google Scholar]
- Atasheva, S.; Wang, E.; Adams, A.P.; Plante, K.S.; Ni, S.; Taylor, K.; Miller, M.E.; Frolov, I.; Weaver, S.C. Chimeric alphavirus vaccine candidates protect mice from intranasal challenge with western equine encephalitis virus. Vaccine 2009, 27, 4309–4319. [Google Scholar] [CrossRef]
- Smith, T.L. The Emerging West Nile Virus: From the Old World to the New. Perspect. Med. Virol. 2006, 16, 133–148. [Google Scholar]
- Monath, T.P.; Vasconcelos, P.F.C. Yellow fever. J. Clin. Virol. 2015, 64, 160–173. [Google Scholar] [CrossRef]
- Vasudevan, J.; Skandhan, A.; Skandhan, A.K.P.; Balakrishnan, S.; Skandhand, K.P. Zika virus. Rev. Med. Microbiol. 2018, 29, 43–50. [Google Scholar] [CrossRef]
- OIE World Organization for Animal Health. Technical Disease Cards—African Horse Sickness. 2013. Available online: http://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/AFRICAN_HORSE_SICKNESS.pdf (accessed on 15 December 2018).
- Mellor, P.S.; Wittmann, E.J. Bluetongue Virus in the Mediterranean Basin 1998–2001. Vet. J. 2002, 164, 20–37. [Google Scholar] [CrossRef]
- Ganter, M. Bluetongue disease—Global overview and future risks. Small Rumin. Res. 2014, 118, 79–85. [Google Scholar] [CrossRef]
- OIE World Organization for Animal Health. Technical Disease Cards—Epizootic Haemorrhagic Disease. 2009. Available online: http://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/EPIZOOTIC_HEAMORRHAGIC_DISEASE.pdf (accessed on 15 December 2018).
- Kara, P.D.; Mather, A.S.; Pretorius, A.; Chetty, T.; Babiuk, S.; Wallace, D.B. Characterisation of putative immunomodulatory gene knockouts of lumpy skin disease virus in cattle towards an improved vaccine. Vaccine 2018, 36, 4708–4715. [Google Scholar] [CrossRef] [PubMed]
- Meyers, A.; Tatu, U. Schmallenberg virus. Resonance 2014, 19, 814–820. [Google Scholar] [CrossRef]
- Almeida-Filho, R.; Miranda, F.P. Mega capture of the Rio Negro and formation of the Anavilhanas Archipelago, Central Amazônia, Brazil: Evidences in an SRTM digital elevation model. Remote Sens. Environ. 2007, 110, 387–392. [Google Scholar] [CrossRef]
- OIE World Organization for Animal Health. Technical Disease Cards—Trypanosoma evansi Infections (Including Surra). 2013. Available online: http://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/TRYPANO_EVANSI.pdf (accessed on 15 December 2018).
- Khasnis, A.A.; Nettleman, M.D. Global warming and infectious disease. Arch. Med. Res. 2005, 36, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Guerrant, R.L.; Blackwood, B.L. Perspective threats to global health and survival: The growing crises of tropical infectious diseases-our “unfinished agenda”. Clin. Infect. Dis. 1999, 28, 966–986. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.-J.; Brook, B.W.; Whelan, P.I.; Cleland, S.; Bradshaw, C.J.A. Endogenous and exogenous factors controlling temporal abundance patterns of tropical mosquitoes. Ecol. Appl. 2008, 18, 2028–2040. [Google Scholar] [CrossRef]
- Rochlin, I.; Gaugler, R.; Williges, E.; Farajollahi, A. The rise of the invasives and decline of the natives: Insights revealed from adult populations of container-inhabiting Aedes mosquitoes (Diptera: Culicidae) in temperate North America. Biol. Invasions 2013, 15, 991–1003. [Google Scholar] [CrossRef]
- Tun-Lin, W.; Burkot, T.R.; Kay, B.H. Effects of temperature and larval diet on development rates and survival of the dengue vector Aedes aegypti in north Queensland, Australia. Med. Vet. Entomol. 2000, 14, 31–37. [Google Scholar] [CrossRef]
- Culler, L.E.; Ayres, M.P.; Virginia, R.A. In a warmer Arctic, mosquitoes avoid increased mortality from predators by growing faster. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151549. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, M.J.; Licht, M.C.; Lehmann, T. Evidence for a recent population expansion in the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol. Biol. Evol. 2001, 18, 1353–1364. [Google Scholar] [CrossRef]
- Onyabe, D.Y.; Conn, J.E. Population genetic structure of the malaria mosquito Anopheles arabiensis across Nigeria suggests range expansion. Mol. Ecol. 2001, 10, 2577–2591. [Google Scholar] [CrossRef]
- Crowl, T.A.; Crist, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Costantini, C.; Ayala, D.; Guelbeogo, W.M.; Pombi, M.; Some, C.Y.; Bassole, I.H.N.; Ose, K.; Fotsing, J.M.; Sagnon, N.; Fontenille, D.; et al. Living at the edge: Biogeographic patterns of habitat segregation conform to speciation by niche expansion in anopheles gambiae. BMC Ecol. 2009, 9, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Pedro, P.M.; Sallum, M.A.M. Spatial expansion and population structure of the neotropical malaria vector, Anopheles darlingi (Diptera: Culicidae). Biol. J. Linn. Soc. 2009, 97, 854–866. [Google Scholar] [CrossRef] [Green Version]
- Medley, K.A. Niche shifts during the global invasion of the Asian tiger mosquito, Aedes albopictus Skuse (Culicidae), revealed by reciprocal distribution models. Glob. Ecol. Biogeogr. 2010, 19, 122–133. [Google Scholar] [CrossRef]
- Hongoh, V.; Berrang-Ford, L.; Scott, M.E.; Lindsay, L.R. Expanding geographical distribution of the mosquito, Culex pipiens, in Canada under climate change. Appl. Geogr. 2012, 33, 53–62. [Google Scholar] [CrossRef]
- Guagliardo, S.A.; Barboza, J.L.; Morrison, A.C.; Astete, H.; Vazquez-Prokopec, G.; Kitron, U. Patterns of geographic expansion of Aedes aegypti in the Peruvian Amazon. PLoS Negl. Trop. Dis. 2014, 8, e3033. [Google Scholar] [CrossRef]
- Weaver, S.C. Urbanization and geographic expansion of zoonotic arboviral diseases: Mechanisms and potential strategies for prevention. Trends Microbiol. 2013, 21, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Leta, S.; Beyene, T.J.; De Clercq, E.M.; Amenu, K.; Kraemer, M.U.G.; Revie, C.W. Global risk mapping for major diseases transmitted by Aedes aegypti and Aedes albopictus. Int. J. Infect. Dis. 2018, 67, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.M. Risks to the Americas associated with the continued expansion of chikungunya virus. J. Gen. Virol. 2015, 96, 1–5. [Google Scholar] [CrossRef]
- Benelli, G.; Mehlhorn, H. Declining malaria, rising of dengue and Zika virus: Insights for mosquito vector control. Parasitol. Res. 2016, 115, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Roth, D.; Henry, B.; Mak, S.; Fraser, M.; Taylor, M.; Li, M.; Cooper, K.; Furnell, A.; Wong, Q.; Morshed, M. West Nile Virus range expansion into British Columbia. Emerg. Infect. Dis. 2010, 16, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Benedict, M.Q.; Levine, R.S.; Hawley, W.A.; Lounibos, L.P. Spread of the tiger: Global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007, 7, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M. A mosquito goes global. Science 2008, 320, 864–866. [Google Scholar] [CrossRef]
- Tatem, A.J.; Hay, S.I.; Rogers, D.J. Global traffic and disease vector dispersal. Proc. Natl. Acad. Sci. USA 2006, 103, 6242–6247. [Google Scholar] [CrossRef] [Green Version]
- Hawley, W.A.; Reiter, P.; Copeland, R.S.; Pumpuni, C.B.; Craig, G.B. Aedes albopictus in North America: Probable introduction in used tires from Northern Asia. Science 1987, 236, 1114–1116. [Google Scholar] [CrossRef]
- Reiter, P.; Sprenger, D. The used tire trade: A mechanism for the worldwide dispersal of container breeding mosquitoes. J. Am. Mosq. Control Assoc. 1987, 3, 494–501. [Google Scholar]
- Juliano, S.A.; Philip Lounibos, L. Ecology of invasive mosquitoes: Effects on resident species and on human health. Ecol. Lett. 2005, 8, 558–574. [Google Scholar] [CrossRef]
- Brown, J.E.; Evans, B.R.; Zheng, W.; Obas, V.; Barrera-Martinez, L.; Egizi, A.; Zhao, H.; Caccone, A.; Powell, J.R. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution 2014, 68, 514–525. [Google Scholar] [CrossRef]
- Kraemer, M.U.G.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.N.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. Albopictus. eLife 2015, 4, 1–18. [Google Scholar] [CrossRef]
- Taylor, C.E.; Toure, Y.T.; Coluzzi, M.; Petrarca, V. Effective population size and persistence of Anopheles arabiensis during the dry season in West Africa. Med. Vet. Entomol. 1993, 7, 351–357. [Google Scholar] [CrossRef]
- Toure, Y.T.; Traore, S.F.; Sankare, O.; Sow, M.Y.; Coulibaly, A.; Esposito, F.; Petrarca, V. Perennial transmission of malaria by the Anopheles gambiae complex in a North Sudan Savanna area of Mali. Med. Vet. Entomol. 1996, 10, 197–199. [Google Scholar] [CrossRef]
- Simard, F.; Lehmann, T.; Lemasson, J.J.; Diatta, M.; Fontenille, D. Persistence of Anopheles arabiensis during the severe dry season conditions in Senegal: An indirect approach using microsatellite loci. Insect Mol. Biol. 2000, 9, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Tonnang, H.E.Z.; Kangalawe, R.Y.M.; Yanda, P.Z. Predicting and mapping malaria under climate change scenarios: The potential redistribution of malaria vectors in Africa. Malar. J. 2010, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Alimi, T.O.; Fuller, D.O.; Qualls, W.A.; Herrera, S.V.; Arevalo-Herrera, M.; Quinones, M.L.; Lacerda, M.V.G.; Beier, J.C. Predicting potential ranges of primary malaria vectors and malaria in northern South America based on projected changes in climate, land cover and human population. Parasites Vectors 2015, 8, 1–16. [Google Scholar] [CrossRef]
- Ciota, A.T.; Kramer, L.D. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 2013, 5, 3021–3047. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.M.; Burkett-Cadena, N.D.; Eubanks, M.D. Distribution expansion of Culex coronator in Alabama. J. Am. Mosq. Control Assoc. 2008, 24, 585–587. [Google Scholar] [CrossRef]
- Connelly, C.R.; Alto, B.W.; O’Meara, G.F. The spread of Culex coronator (Diptera: Culicidae) throughout Florida. J. Vector Ecol. 2016, 41, 195–199. [Google Scholar] [CrossRef] [Green Version]
- Akaratovic, K.I.; Kiser, J.P. First record of culex coronator in Virginia, with notes on its rapid dispersal, trapping methods, and biology. J. Am. Mosq. Control Assoc. 2017, 33, 225–228. [Google Scholar] [CrossRef]
- Samy, A.M.; Elaagip, A.H.; Kenawy, M.A.; Ayres, C.F.J.; Peterson, A.T.; Soliman, D.E. Climate change influences on the global potential distribution of the mosquito Culex quinquefasciatus, vector of West Nile virus and lymphatic filariasis. PLoS ONE 2016, 11, e0163863. [Google Scholar] [CrossRef]
- Ramasamy, R.; Surendran, S.N. Global climate change and its potential impact on disease transmission by salinity-tolerant mosquito vectors in coastal zones. Front. Physiol. 2012, 3, 1–14. [Google Scholar] [CrossRef]
- Ramasamy, R.; Surendran, S.N.; Jude, P.J.; Dharshini, S.; Vinobaba, M. Larval development of Aedes aegypti and Aedes albopictus in peri-urban brackish water and its implications for transmission of arboviral diseases. PLoS Negl. Trop. Dis. 2011, 5, e1369. [Google Scholar] [CrossRef] [PubMed]
- Pfennig, K.S.; Kelly, A.L.; Pierce, A.A. Hybridization as a facilitator of species range expansion. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, J.R.; Petrarca, V.; della Torre, A.; Caccone, A.; Coluzzi, M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia 1999, 41, 101–113. [Google Scholar]
- Besansky, N.J.; Krzywinski, J.; Lehmann, T.; Simard, F.; Kern, M.; Mukabayire, O.; Fontenille, D.; Toure, Y.; Sagnon, N. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 2003, 100, 10818–10823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beebe, N.W.; Cooper, R.D.; Mottram, P.; Sweeney, A.W. Australia’s dengue risk driven by human adaptation to climate change. PLoS Negl. Trop. Dis. 2009, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O. The global distribution and burden of dengue. Nature 2013, 496, 504. [Google Scholar] [CrossRef] [PubMed]
- Imai, N.; Dorigatti, I.; Cauchemez, S.; Ferguson, N.M. Estimating dengue transmission intensity from sero-prevalence surveys in multiple countries. PLoS Negl. Trop. Dis. 2015, 9, e0003719. [Google Scholar] [CrossRef]
- Aubry, M.; Teissier, A.; Huart, M.; Merceron, S.; Vanhomwegen, J.; Roche, C.; Vial, A.-L.; Teururai, S.; Sicard, S.; Paulous, S. Zika virus seroprevalence, French Polynesia, 2014–2015. Emerg. Infect. Dis. 2017, 23, 669. [Google Scholar] [CrossRef]
- Netto, E.M.; Moreira-Soto, A.; Pedroso, C.; Höser, C.; Funk, S.; Kucharski, A.J.; Rockstroh, A.; Kümmerer, B.M.; Sampaio, G.S.; Luz, E.; et al. High Zika Virus Seroprevalence in Salvador, Northeastern Brazil Limits the Potential for Further Outbreaks. mBio 2017, 8, e01390-17. [Google Scholar] [CrossRef]
- Lourenço, J.; Monteiro, M.; Tomás, T.; Monteiro Rodrigues, J.; Pybus, O.; Rodrigues Faria, N. Epidemiology of the zika virus outbreak in the Cabo Verde Islands, West Africa. PLoS Curr. 2018, 1, 5866102. [Google Scholar] [CrossRef]
- Kucharski, A.J.; Funk, S.; Eggo, R.M.; Mallet, H.-P.; Edmunds, W.J.; Nilles, E.J. Transmission dynamics of zika virus in island populations: A modelling analysis of the 2013–14 French Polynesia Outbreak. PLoS Negl. Trop. Dis. 2016, 10, e0004726. [Google Scholar] [CrossRef] [PubMed]
- Hales, S.; de Wet, N.; Maindonald, J.; Woodward, A. Potential effect of population and climate changes on global distribution of dengue fever: An empirical model. Lancet 2002, 360, 830–834. [Google Scholar] [CrossRef]
- Chretien, J.-P.; Swedlow, D.; Eckstrand, I.; George, D.; Johansson, M.; Huffman, R.; Hebbeler, A. Advancing epidemic prediction and forecasting: A new US Government initiative. Online J. Public Health Inform. 2015, 7, e13. [Google Scholar] [CrossRef]
- Stewart-Ibarra, A.M.; Lowe, R. Climate and non-climate drivers of dengue epidemics in southern coastal Ecuador. Am. J. Trop. Med. Hyg. 2013, 88, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Stewart-Ibarra, A.M.; Petrova, D.; García-Díez, M.; Borbor-Cordova, M.J.; Mejía, R.; Regato, M.; Rodó, X. Climate services for health: Predicting the evolution of the 2016 dengue season in Machala, Ecuador. Lancet Planet. Heal. 2017, 1, e142–e151. [Google Scholar] [CrossRef]
- Liyanage, P.; Tissera, H.; Sewe, M.; Quam, M.; Amarasinghe, A.; Palihawadana, P.; Wilder-Smith, A.; Louis, V.; Tozan, Y.; Rocklöv, J. A spatial hierarchical analysis of the temporal influences of the El Niño-Southern Oscillation and weather on dengue in Kalutara District, Sri Lanka. Int. J. Environ. Res. Public Health 2016, 13, 1087. [Google Scholar] [CrossRef] [PubMed]
- Teurlai, M.; Menkès, C.E.; Cavarero, V.; Degallier, N.; Descloux, E.; Grangeon, J.-P.; Guillaumot, L.; Libourel, T.; Lucio, P.S.; Mathieu-Daudé, F.; et al. Socio-economic and climate factors associated with dengue fever spatial heterogeneity: A worked example in New Caledonia. PLoS Negl. Trop. Dis. 2015, 9, e0004211. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, T.; Liu, K.; Xia, Y.; Lu, Y.; Jing, Q.; Yang, Z.; Hu, W.; Lu, J. Developing a time series predictive model for dengue in Zhongshan, China based on weather and Guangzhou dengue surveillance data. PLoS Negl. Trop. Dis. 2016, 10, e0004473. [Google Scholar] [CrossRef]
- Ramachandran, V.G.; Roy, P.; Das, S.; Mogha, N.S.; Bansal, A.K. Empirical model for calculating dengue incidence using temperature, rainfall and relative humidity: A 19-year retrospective analysis in East Delhi, India. Epidemiol. Health 2016, 38, e2016052. [Google Scholar] [CrossRef]
- Lowe, R.; Coelho, C.A.; Barcellos, C.; Carvalho, M.S.; Catão, R.D.C.; Coelho, G.E.; Ramalho, W.M.; Bailey, T.C.; Stephenson, D.B.; Rodó, X. Evaluating probabilistic dengue risk forecasts from a prototype early warning system for Brazil. eLife 2016, 5, e11285. [Google Scholar] [CrossRef]
- Adde, A.; Roucou, P.; Mangeas, M.; Ardillon, V.; Desenclos, J.-C.; Rousset, D.; Girod, R.; Briolant, S.; Quenel, P.; Flamand, C. Predicting dengue fever outbreaks in French Guiana using climate indicators. PLoS Negl. Trop. Dis. 2016, 10, e0004681. [Google Scholar] [CrossRef]
- Morin, C.W.; Comrie, A.C.; Ernst, K. Climate and dengue transmission: Evidence and implications. Environ. Health Perspect. 2013, 121, 1264–1272. [Google Scholar] [CrossRef]
- Reiner, R.C.; Perkins, T.A.; Barker, C.M.; Niu, T.; Chaves, L.F.; Ellis, A.M.; George, D.B.; Le Menach, A.; Pulliam, J.R.C.; Bisanzio, D.; et al. A systematic review of mathematical models of mosquito-borne pathogen transmission: 1970–2010. J. R. Soc. Interface 2013, 10, 20120921. [Google Scholar] [CrossRef]
- Braga, C.; Luna, C.F.; Martelli, C.M.; de Souza, W.V.; Cordeiro, M.T.; Alexander, N.; de Albuquerque, M.F.P.M.; Júnior, J.C.S.; Marques, E.T. Seroprevalence and risk factors for dengue infection in socio-economically distinct areas of Recife, Brazil. Acta Trop. 2010, 113, 234–240. [Google Scholar] [CrossRef]
- Akter, R.; Hu, W.; Naish, S.; Banu, S.; Tong, S. Joint effects of climate variability and socioecological factors on dengue transmission: Epidemiological evidence. Trop. Med. Int. Heal. 2017, 22, 656–669. [Google Scholar] [CrossRef]
- Mulligan, K.; Dixon, J.; Joanna Sinn, C.-L.; Elliott, S.J. Is dengue a disease of poverty? A systematic review. Pathog. Glob. Health 2015, 109, 10–18. [Google Scholar] [CrossRef]
- Louis, V.R.; Phalkey, R.; Horstick, O.; Ratanawong, P.; Wilder-Smith, A.; Tozan, Y.; Dambach, P. Modeling tools for dengue risk mapping—A systematic review. Int. J. Health Geogr. 2014, 13, 50. [Google Scholar] [CrossRef]
- Buczak, A.L.; Koshute, P.T.; Babin, S.M.; Feighner, B.H.; Lewis, S.H. A data-driven epidemiological prediction method for dengue outbreaks using local and remote sensing data. BMC Med. Inform. Decis. Mak. 2012, 12, 124. [Google Scholar] [CrossRef]
- Machault, V.; Yébakima, A.; Etienne, M.; Vignolles, C.; Palany, P.; Tourre, Y.; Guérécheau, M.; Lacaux, J.-P. Mapping entomological dengue risk levels in Martinique using high-resolution remote-sensing environmental data. ISPRS Int. J. Geo-Inf. 2014, 3, 1352–1371. [Google Scholar] [CrossRef]
- Kalluri, S.; Gilruth, P.; Rogers, D.; Szczur, M. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: A review. PLoS Pathog. 2007, 3, e116. [Google Scholar] [CrossRef]
- Tian, H.; Huang, S.; Zhou, S.; Bi, P.; Yang, Z.; Li, X.; Chen, L.; Cazelles, B.; Yang, J.; Luo, L.; et al. Surface water areas significantly impacted 2014 dengue outbreaks in Guangzhou, China. Environ. Res. 2016, 150, 299–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.; Bi, D.; Xie, G.; Jin, Y.; Huang, Y.; Lin, B.; An, X.; Feng, D.; Tong, Y. Dynamic forecasting of zika epidemics using Google Trends. PLoS ONE 2017, 12, e0165085. [Google Scholar] [CrossRef]
- Gluskin, R.T.; Johansson, M.A.; Santillana, M.; Brownstein, J.S. Evaluation of internet-based dengue query data: Google Dengue Trends. PLoS Negl. Trop. Dis. 2014, 8, e2713. [Google Scholar] [CrossRef]
- Moran, K.R.; Fairchild, G.; Generous, N.; Hickmann, K.; Osthus, D.; Priedhorsky, R.; Hyman, J.; Del Valle, S.Y. Epidemic forecasting is messier than weather forecasting: The role of human behavior and internet data streams in epidemic forecast. J. Infect. Dis. 2016, 214, S404–S408. [Google Scholar] [CrossRef]
- Yang, S.; Kou, S.C.; Lu, F.; Brownstein, J.S.; Brooke, N.; Santillana, M. Advances in using Internet searches to track dengue. PLOS Comput. Biol. 2017, 13, e1005607. [Google Scholar] [CrossRef]
- de Almeida Marques-Toledo, C.; Degener, C.M.; Vinhal, L.; Coelho, G.; Meira, W.; Codeço, C.T.; Teixeira, M.M. Dengue prediction by the web: Tweets are a useful tool for estimating and forecasting Dengue at country and city level. PLoS Negl. Trop. Dis. 2017, 11, e0005729. [Google Scholar] [CrossRef]
- Rivera, R. A dynamic linear model to forecast hotel registrations in Puerto Rico using Google Trends data. Tour. Manag. 2016, 57, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Strauss, R.A.; Castro, J.S.; Reintjes, R.; Torres, J.R. Google dengue trends: An indicator of epidemic behavior. The Venezuelan Case. Int. J. Med. Inform. 2017, 104, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Althouse, B.M.; Ng, Y.Y.; Cummings, D.A.T. Prediction of dengue incidence using search query surveillance. PLoS Negl. Trop. Dis. 2011, 5, e1258. [Google Scholar] [CrossRef]
- Lowe, R.; Barcellos, C.; Coelho, C.A.S.; Bailey, T.C.; Coelho, G.E.; Graham, R.; Jupp, T.; Ramalho, W.M.; Carvalho, M.S.; Stephenson, D.B.; et al. Dengue outlook for the World Cup in Brazil: An early warning model framework driven by real-time seasonal climate forecasts. Lancet Infect. Dis. 2014, 14, 619–626. [Google Scholar] [CrossRef]
- Racloz, V.; Ramsey, R.; Tong, S.; Hu, W. Surveillance of dengue fever virus: A review of epidemiological models and early warning systems. PLoS Negl. Trop. Dis. 2012, 6, e1648. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Bailey, T.C.; Stephenson, D.B.; Graham, R.J.; Coelho, C.A.S.; Sá Carvalho, M.; Barcellos, C. Spatio-temporal modelling of climate-sensitive disease risk: Towards an early warning system for dengue in Brazil. Comput. Geosci. 2011, 37, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.-L.; Yang, S.-J.; Yen, H.-J.; Christakos, G. A spatio-temporal climate-based model of early dengue fever warning in southern Taiwan. Stoch. Environ. Res. Risk Assess. 2011, 25, 485–494. [Google Scholar] [CrossRef]
- Salje, H.; Lessler, J.; Endy, T.P.; Curriero, F.C.; Gibbons, R.V.; Nisalak, A.; Nimmannitya, S.; Kalayanarooj, S.; Jarman, R.G.; Thomas, S.J.; et al. Revealing the microscale spatial signature of dengue transmission and immunity in an urban population. Proc. Natl. Acad. Sci. USA 2012, 109, 9535–9538. [Google Scholar] [CrossRef] [Green Version]
- McGough, S.F.; Brownstein, J.S.; Hawkins, J.B.; Santillana, M. Forecasting zika incidence in the 2016 latin america outbreak combining traditional disease surveillance with search, social media, and news report data. PLoS Negl. Trop. Dis. 2017, 11, e0005295. [Google Scholar] [CrossRef]
- Ashby, J.; Moreno-Madriñán, M.; Yiannoutsos, C.; Stanforth, A. Niche modeling of dengue fever using remotely sensed environmental factors and boosted regression trees. Remote Sens. 2017, 9, 328. [Google Scholar] [CrossRef]
- Peterson, A.T.; Martínez-Campos, C.; Nakazawa, Y.; Martínez-Meyer, E. Time-specific ecological niche modeling predicts spatial dynamics of vector insects and human dengue cases. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Kraemer, M.U.; Brady, O.J.; Pigott, D.M.; Shearer, F.M.; Weiss, D.J.; Golding, N.; Ruktanonchai, C.W.; Gething, P.W.; Cohn, E.; et al. Mapping global environmental suitability for Zika virus. eLife 2016, 5, e15272. [Google Scholar] [CrossRef]
- Nsoesie, E.O.; Kraemer, M.U.G.; Golding, N.; Pigott, D.M.; Brady, O.J.; Moyes, C.L.; Johansson, M.A.; Gething, P.W.; Velayudhan, R.; Khan, K.; et al. Global distribution and environmental suitability for chikungunya virus, 1952 to 2015. Eurosurveillance 2016, 21, 30234. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Stige, L.C.; Chan, K.-S.; Zhou, J.; Yang, J.; Sang, S.; Wang, M.; Yang, Z.; Yan, Z.; Jiang, T.; et al. Climate variation drives dengue dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, 113–118. [Google Scholar] [CrossRef]
- Manore, C.A.; Davis, J.K.; Christofferson, R.C.; Wesson, D.M.; Hyman, J.M.; Mores, C.N. Towards an early warning system for forecasting human West Nile virus incidence. PLoS Curr. 2014, 6, 25914857. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, K.; Chinazzi, M.; y Piontti, A.P.; Dean, N.E.; Rojas, D.P.; Merler, S.; Mistry, D.; Poletti, P.; Rossi, L. Spread of Zika virus in the Americas. Proc. Natl. Acad. Sci. USA 2017, 114, E4334–E4343. [Google Scholar] [CrossRef] [Green Version]
- Ewing, D.A.; Cobbold, C.A.; Purse, B.V.; Nunn, M.A.; White, S.M. Modelling the effect of temperature on the seasonal population dynamics of temperate mosquitoes. J. Theor. Biol. 2016, 400, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Lunde, T.M.; Bayoh, M.N.; Lindtjørn, B. How malaria models relate temperature to malaria transmission. Parasit. Vectors 2013, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Magori, K.; Legros, M.; Puente, M.E.; Focks, D.A.; Scott, T.W.; Lloyd, A.L.; Gould, F. Skeeter Buster: A stochastic, spatially explicit modeling tool for studying Aedes aegypti population replacement and population suppression strategies. PLoS Negl. Trop. Dis. 2009, 3, e508. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Legros, M.; Gould, F.; Lloyd, A.L. Understanding uncertainties in model-based predictions of Aedes aegypti population dynamics. PLoS Negl. Trop. Dis. 2010, 4, e830. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Ahumada, J.A.; Chaves, L.F.; Rodo, X.; Bouma, M. Malaria resurgence in the East African highlands: Temperature trends revisited. Proc. Natl. Acad. Sci. USA 2006, 103, 5829–5834. [Google Scholar] [CrossRef] [Green Version]
- Erguler, K.; Smith-Unna, S.E.; Waldock, J.; Proestos, Y.; Christophides, G.K.; Lelieveld, J.; Parham, P.E. Large-scale modelling of the environmentally-driven population dynamics of temperate Aedes albopictus (Skuse). PLoS ONE 2016, 11, e0149282. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Pigott, D.M.; Golding, N.; Kraemer, M.U.G.; Scott, T.W.; Wint, G.R.W.; Smith, D.L.; Hay, S.I. The many projected futures of dengue. Nat. Rev. Microbiol. 2015, 13, 230–239. [Google Scholar] [CrossRef]
- Okuneye, K.; Gumel, A.B. Analysis of a temperature- and rainfall-dependent model for malaria transmission dynamics. Math. Biosci. 2017, 287, 72–92. [Google Scholar] [CrossRef]
- Parham, P.E.; Michael, E. Modeling the effects of weather and climate change on Malaria transmission. Environ. Health Perspect. 2010, 118, 620–626. [Google Scholar] [CrossRef]
- Abdelrazec, A.; Gumel, A.B. Mathematical assessment of the role of temperature and rainfall on mosquito population dynamics. J. Math. Biol. 2017, 74, 1351–1395. [Google Scholar] [CrossRef]
- Tompkins, A.M.; Ermert, V. A regional-scale, high resolution dynamical malaria model that accounts for population density, climate and surface hydrology. Malar. J. 2013, 12, 65. [Google Scholar] [CrossRef]
- Bomblies, A.; Duchemin, J.-B.; Eltahir, E.A.B. Hydrology of malaria: Model development and application to a Sahelian village. Water Resour. Res. 2008, 44. [Google Scholar] [CrossRef] [Green Version]
- Soti, V.; Tran, A.; Degenne, P.; Chevalier, V.; Lo Seen, D.; Thiongane, Y.; Diallo, M.; Guégan, J.-F.; Fontenille, D. Combining hydrology and mosquito population models to identify the drivers of rift valley fever emergence in semi-arid regions of West Africa. PLoS Negl. Trop. Dis. 2012, 6, e1795. [Google Scholar] [CrossRef]
- Little, E.; Bajwa, W.; Shaman, J. Local environmental and meteorological conditions influencing the invasive mosquito Ae. albopictus and arbovirus transmission risk in New York City. PLoS Negl. Trop. Dis. 2017, 11, e0005828. [Google Scholar] [CrossRef]
- Colón-González, F.J.; Fezzi, C.; Lake, I.R.; Hunter, P.R. The effects of weather and climate change on dengue. PLoS Negl. Trop. Dis. 2013, 7, e2503. [Google Scholar] [CrossRef]
- Naish, S.; Dale, P.; Mackenzie, J.S.; McBride, J.; Mengersen, K.; Tong, S.L. Climate change and dengue: A critical and systematic review of quantitative modelling approaches. BMC Infect. Dis. 2014, 14, 167. [Google Scholar] [CrossRef]
- Johnson, L.R.; Gramacy, R.B.; Cohen, J.; Mordecai, E.; Murdock, C.; Rohr, J.; Ryan, S.J.; Stewart-Ibarra, A.M.; Weikel, D. Phenomenological forecasting of disease incidence using heteroskedastic Gaussian processes: A dengue case study. Ann. Appl. Stat. 2018, 12, 27–66. [Google Scholar] [CrossRef] [Green Version]
- Martin, V.; Chevalier, V.Ã.; Ceccato, P.N.; Anyamba, A.; De Simone, L.; Lubroth, J.; Domenech, J. The impact of climate change on the epidemiology and control of Rift Valley fever. Rev. Sci. Tech. 2008, 27, 413–426. [Google Scholar] [CrossRef]
- Sallam, M.; Fizer, C.; Pilant, A.; Whung, P. Systematic review: Land cover, meteorological, and socioeconomic determinants of Aedes mosquito habitat for risk mapping. Int. J. Environ. Res. Public Health 2017, 14, 1230. [Google Scholar] [CrossRef] [PubMed]
- McMahon, B.H.; Manore, C.A.; Hyman, J.M.; LaBute, M.X.; Fair, J.M. Coupling vector-host dynamics with weather geography and mitigation measures to model rift valley fever in Africa. Math. Model. Nat. Phenom. 2014, 9, 161–177. [Google Scholar] [CrossRef]
- Manore, C.A.; Beechler, B.R. Inter-epidemic and between-season persistence of rift valley fever: Vertical transmission or cryptic cycling? Transbound. Emerg. Dis. 2015, 62, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, Á.G.; Thomson, M.C.; Stewart-Ibarra, A.M.; Vecchi, G.A.; Chourio, X.; Nájera, P.; Moran, Z.; Yang, X. Could the recent zika epidemic have been predicted? Front. Microbiol. 2017, 8, 1291. [Google Scholar] [CrossRef]
- Zouache, K.; Fontaine, A.; Vega-Rua, A.; Mousson, L.; Thiberge, J.-M.; Lourenco-De-Oliveira, R.; Caro, V.; Lambrechts, L.; Failloux, A.-B. Three-way interactions between mosquito population, viral strain and temperature underlying chikungunya virus transmission potential. Proc. R. Soc. B Biol. Sci. 2014, 281, 20141078. [Google Scholar] [CrossRef] [PubMed]
- Kassa, S.M.; Ouhinou, A. The impact of self-protective measures in the optimal interventions for controlling infectious diseases of human population. J. Math. Biol. 2015, 70, 213–236. [Google Scholar] [CrossRef]
- Agusto, F.B.; Gumel, A.B.; Parham, P.E. Qualitative assessment of the role of temperature variations on malaria transmission dynamics. J. Biol. Syst. 2015, 23, 1550030. [Google Scholar] [CrossRef]
- Sellers, W.D. A global climatic model based on the energy balance of the Earth-atmosphere system. J. Appl. Meteorol. 1969, 8, 392–400. [Google Scholar] [CrossRef]
- Bryan, K. Climate and the ocean circulation. Mon. Weather Rev 1969, 97, 806–827. [Google Scholar] [CrossRef]
- Manabe, S. Climate and the ocean circulation: I. The atmospheric circulation and the hydrology of the earth’s surface. Mon. Weather Rev. 1969, 97, 739–774. [Google Scholar] [CrossRef]
- Hayhoe, K.; Edmonds, J.; Kopp, R.E.; LeGrande, A.N.; Sanderson, B.M.; Wehner, M.F.; Wuebbles, D.J. Climate Models, Scenarios, and Projections. In Climate Science Special Report: Fourth National Climate Assessment, Volume I.; U.S. Global Change Research Program: Washington, DC, USA, 2017. [Google Scholar]
- Flato, G.M. Earth system models: An overview. Wiley Interdiscip. Rev. Clim. Chang. 2011, 2, 783–800. [Google Scholar] [CrossRef]
- Calvin, K.; Bond-Lamberty, B. Integrated human-earth system modeling—State of the science and future directions. Environ. Res. Lett. 2018, 13, 063006. [Google Scholar] [CrossRef]
- Boyce, R.; Reyes, R.; Matte, M.; Ntaro, M.; Mulogo, E.; Metlay, J.P.; Band, L.; Siedner, M.J. Severe flooding and malaria transmission in the western Ugandan Highlands: Implications for disease control in an era of global climate change. J. Infect. Dis. 2016, 214, 1403–1410. [Google Scholar] [CrossRef]
- Paull, S.H.; Horton, D.E.; Ashfaq, M.; Rastogi, D.; Kramer, L.D.; Diffenbaugh, N.S.; Kilpatrick, A.M. Drought and immunity determine the intensity of West Nile virus epidemics and climate change impacts. Proc. R. Soc. B Biol. Sci. 2017, 284, 20162078. [Google Scholar] [CrossRef] [Green Version]
- Caminade, C.; Turner, J.; Metelmann, S.; Hesson, J.C.; Blagrove, M.S.C.; Solomon, T.; Morse, A.P.; Baylis, M. Global risk model for vector-borne transmission of Zika virus reveals the role of El Niño 2015. Proc. Natl. Acad. Sci. USA 2017, 114, 119–124. [Google Scholar] [CrossRef]
- Wesolowski, A.; Eagle, N.; Tatem, A.J.; Smith, D.L.; Noor, A.M.; Snow, R.W.; Buckee, C.O. Quantifying the impact of human mobility on malaria. Science 2012, 338, 267–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deville, P.; Linard, C.; Martin, S.; Gilbert, M.; Stevens, F.R.; Gaughan, A.E.; Blondel, V.D.; Tatem, A.J. Dynamic population mapping using mobile phone data. Proc. Natl. Acad. Sci. USA 2014, 111, 15888–15893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaDeau, S.L.; Glass, G.E.; Hobbs, N.T.; Latimer, A.; Ostfeld, R.S. Data–model fusion to better understand emerging pathogens and improve infectious disease forecasting. Ecol. Appl. 2011, 21, 1443–1460. [Google Scholar] [CrossRef] [PubMed]
- Ostfeld, R.; Glass, G.; Keesing, F. Spatial epidemiology: An emerging (or re-emerging) discipline. Trends Ecol. Evol. 2005, 20, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Kitron, U.; Clennon, J.A.; Cecere, M.C.; Gürtler, R.E.; King, C.H.; Vazquez-Prokopec, G. Upscale or downscale: Applications of fine scale remotely sensed data to Chagas disease in Argentina and schistosomiasis in Kenya. Geospat. Health 2006, 1, 49. [Google Scholar] [CrossRef]
- Glass, G.E. Rainy with a chance of plague: Forecasting disease outbreaks from satellites. Future Virol. 2007, 2, 225–229. [Google Scholar] [CrossRef]
- Hobbs, N.T.; Hilborn, R. Alternatives to statistical hypothesis testing in ecology: A guide to self teaching. Ecol. Appl. 2006, 16, 5–19. [Google Scholar] [CrossRef]
- Bjørnstad, O.N.; Grenfell, B.T. Noisy clockwork: Time series analysis of population fluctuations in animals. Science 2001, 293, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Ball, C.; Priya, A.; Braswell, T.; Renzi, R.; Helm, J.; Coffey, L.; Meagher, R. Smart trap for autonomous monitoring of mosquito-borne viruses. In Proceedings of the 2017 Defense Threat Reduction Agency (DTRA) Chemical and Biological Defense Science & Technology (CBD S&T) Conference, Long Beach, CA, USA, 28–30 November 2017. [Google Scholar]
- Polgreen, P.M.; Chen, Y.; Pennock, D.M.; Nelson, F.D. Using internet searches for influenza surveillance. Clin. Infect. Dis. 2008, 47, 1443–1448. [Google Scholar] [CrossRef]
- Ginsberg, J.; Mohebbi, M.H.; Patel, R.S.; Brammer, L.; Smolinski, M.S.; Brilliant, L. Detecting influenza epidemics using search engine query data. Nature 2009, 457, 1012–1014. [Google Scholar] [CrossRef] [PubMed]
- Signorini, A.; Segre, A.M.; Polgreen, P.M. The use of Twitter to track levels of disease activity and public concern in the U.S. during the Influenza A H1N1 pandemic. PLoS ONE 2011, 6, e19467. [Google Scholar] [CrossRef] [PubMed]
- Generous, N.; Fairchild, G.; Deshpande, A.; Del Valle, S.Y.; Priedhorsky, R. Global disease monitoring and forecasting with Wikipedia. PLoS Comput. Biol. 2014, 10, e1003892. [Google Scholar] [CrossRef]
- Ziemann, A.; Fairchild, G.; Conrad, J.; Manore, C.; Parikh, N.; Del Valle, S.; Generous, N. Predicting dengue incidence in Brazil using broad-scale spectral remote sensing imagery. In Proceedings of the IGARSS 2018—2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 2076–2078. [Google Scholar]
- Stein, M.L. Interpolation of Spatial Data; Springer: New York, NY, USA, 1999. [Google Scholar]
- Gelfand, A.E.; Diggle, P.; Guttorp, P.; Fuentes, M. Handbook of Spatial Statistics; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Clark, J.S.; Ferraz, G.; Oguge, N.; Hays, H.; DiCostanzo, J. Hierarchical Bayes for structured, variable populations: From recapture data to life-history prediction. Ecology 2005, 86, 2232–2244. [Google Scholar] [CrossRef]
- Qian, S.S.; Cuffney, T.F.; Alameddine, I.; McMahon, G.; Reckhow, K.H. On the application of multilevel modeling in environmental and ecological studies. Ecology 2010, 91, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladeau, S.L.; Clark, J.S. Elevated CO2 and tree fecundity: The role of tree size, interannual variability, and population heterogeneity. Glob. Chang. Biol. 2006, 12, 822–833. [Google Scholar] [CrossRef]


| Pathogen | Taxon Group | Vectors | Animal Hosts | Range |
|---|---|---|---|---|
| Banna virus | Reoviridae | Mosquitoes (Culex, Anopheles, Aedes) [12] | Pigs, cattle, humans [12] | Southeast Asia, Indonesia [12] |
| Chikungunya virus | Togaviridae [13] | Mosquitoes (Aedes) [13] | Humans [13] | Central America, North America, South America, Oceania, central/southern Africa, southern/eastern Asia, western/central Europe [14] |
| Dengue virus | Flaviviridae | Mosquitoes (primarily Aedes aegypti) [15] | Humans [15] In vitro successful infection of amphibians, mammals, & reptiles [16] | North America, Central America, South America, Oceania, Europe, southern Asia, and Africa [17] |
| Dirofilaria spp. (Dirofilariasis) | Nematode | Mosquitoes (Culex, Aedes, Anopheles, Mansonia) [18] | Dogs, cats, humans [19] | Southern/central/eastern Europe, Middle East, eastern/southeastern/central Asia, southeastern North America [19] |
| Eastern Equine Encephalitis virus | Flaviviridae | Mosquitoes (Culiseta, Culex) [20] | Birds, humans, horses [20] | United States [21] |
| Indiana vesiculovirus | Rhabdoviridae | Sand flies (Phlebotomus, Lutzomyia), mosquitoes (Aedes), black flies (family Simuliidae) [22] | Equids, bovids [22] | North America, South America [22] |
| Japanese Encephalitis virus | Flaviviridae | Mosquitoes (Culex) [17] | Pigs, birds, horses, humans [17] | Western Europe, Russia, southern Asia, Oceania [17] |
| Plasmodium spp. (Malaria) | Protozoan | Mosquitoes (Anopheles) [23] | Humans, birds (carriers) [23] | United States, central/southern Africa, southeast Asia, southern Europe [23,24] |
| Mayaro virus | Togaviridae | Mosquitoes (Haemagogus) [25] | Humans [25] | Northern South America *tourists infected [25] |
| Murray Valley Encephalitis virus | Flaviviridae | Mosquitoes (Culex) [26] | Humans [26] | Australia, Northern Territories [26] |
| O’nyong’nyong virus | Togaviridae | Mosquitoes (Anopheles) [27] | Humans [27] | Western/central Africa [27] |
| Oropouche virus | Peribunyaviridae | Mosquitoes (Coquillettidia) [28] | Humans [29] | Central/northern South America, southeastern Central America [28] |
| Rift Valley Fever virus | Phenuviridae | Mosquitoes (Culex, Aedes, Anopheles, Eretmapodites, Mansonia, Culicoides, Coquillettidia) [30] | Ruminants (reservoir), humans [30], Bats [31] | Africa, southern Middle East [30] |
| Saint Louis Encephalitis virus | Flaviviridae | Mosquitoes (Culex) [32] | Birds [32], humans [33] | South America [33], North America [33] |
| Spondweni virus | Flaviviridae | Mosquitoes (Culex, Aedes) [34] | Humans [34] | Caribbean, southern Africa [34] |
| Trypanosoma brucei (Sleeping Sickness) | Protozoan | Tsetse fly (Glossina) [35] | Wild ungulates, ruminants, equids, dogs, humans [35] | Africa, Central America, South America [35] |
| Usutu virus | Flaviviridae | Mosquitoes (Culex) [36] | Birds, horses, humans [36], bats [37] | Central/southern Africa, central Europe [36] |
| Venezuelan Equine Encephalitis virus | Togaviridae | Mosquitoes (Culex) [38] | Horses, humans [38] | Southern North America, central America, northern/central South America [38] |
| Western Equine Encephalitis virus | Togaviridae | Mosquitoes (Culex) [39] | Birds, humans [39] | Western North America, South America, Cuba [39] |
| West Nile virus | Flaviviridae | Mosquitoes (Culex, Aedes) [40] | Birds (reservoir), equids, humans [40] | Africa, the Middle East, Oceania, North America, Central America, northwestern and southern/central South America, and Europe [40] |
| Yellow Fever virus | Flaviviridae | Mosquitoes (Aedes aegypti, Haemagogus) [41] | Humans [41] | Central America, South America, southern North America, Oceania, Europe, southern Asia, and Africa [17] |
| Zika virus | Flaviviridae | Mosquitoes (Aedes) [42] | Primates, humans [42] | central-western and southern Africa, Oceania, South America, North America, Central America, southern Asia, Europe [17] |
| Pathogen | Taxon Group | Vectors | Animal Hosts | Range |
|---|---|---|---|---|
| African Horse Sickness virus | Reoviridae | Midges (Culicoides) [43] | Equids [43] | Central/southern Africa [43] |
| Bluetongue Disease virus | Reoviridae | Midges (Culicoides) [44] | Ruminants [44] | North America, Central America, South America, Africa, southern Asia, northern Australia, Estonia and Russia, southern and central Europe [45] |
| Epizootic Haemorrhagic Disease virus | Reoviridae | Midges (Culicoides) [46] | Ruminants [46] | North America, Australia, Africa, Asia, and the Mediterranean [46] |
| Lumpy Skin Disease virus | Poxviridae | Mosquitoes (Aedes Anopheles), flies (Musca, Stomoxys, Glossina), and midges (Culicoides) [47] | Cattle [47] | Africa, the Middle East, South-Eastern Europe, Russia [47] |
| Schmallenberg virus | Peribunyaviridae | Midges (Culicoides) [48] | Ruminants [49] | Europe [49] |
| Trypanosoma evansi (Surra) | Protozoan | Hematophagous flies (Tabanus, Musca) [50] | Vampire bats (reservoir), ungulates, ruminants, dogs, cats [50] | Asia, northern Africa, Central America, South America, the Middle East [50] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartlow, A.W.; Manore, C.; Xu, C.; Kaufeld, K.A.; Del Valle, S.; Ziemann, A.; Fairchild, G.; Fair, J.M. Forecasting Zoonotic Infectious Disease Response to Climate Change: Mosquito Vectors and a Changing Environment. Vet. Sci. 2019, 6, 40. https://doi.org/10.3390/vetsci6020040
Bartlow AW, Manore C, Xu C, Kaufeld KA, Del Valle S, Ziemann A, Fairchild G, Fair JM. Forecasting Zoonotic Infectious Disease Response to Climate Change: Mosquito Vectors and a Changing Environment. Veterinary Sciences. 2019; 6(2):40. https://doi.org/10.3390/vetsci6020040
Chicago/Turabian StyleBartlow, Andrew W., Carrie Manore, Chonggang Xu, Kimberly A. Kaufeld, Sara Del Valle, Amanda Ziemann, Geoffrey Fairchild, and Jeanne M. Fair. 2019. "Forecasting Zoonotic Infectious Disease Response to Climate Change: Mosquito Vectors and a Changing Environment" Veterinary Sciences 6, no. 2: 40. https://doi.org/10.3390/vetsci6020040