Histopathologic Characterization of Experimental Peracute SARS-CoV-2 Infection in the Syrian Hamster
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study Approval
2.2. Virus
2.3. Experimental Design
2.4. Histology and Immunohistochemistry
2.5. Virology
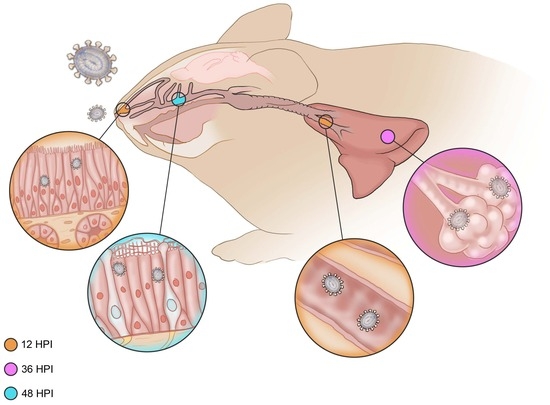
3. Results
3.1. Gross Pathology
3.2. Histopathology
3.2.1. Six Hours Post Inoculation
3.2.2. Twelve Hours Post Inoculation
3.2.3. Thirty-Six Hours Post Inoculation
3.2.4. Forty-Eight Hours Post Inoculation
3.2.5. Seventy-Two Hours Post Inoculation
3.3. Virology
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Carossino, M.; Kenney, D.; O’Connell, A.K.; Montanaro, P.; Tseng, A.E.; Gertje, H.P.; Grosz, K.A.; Ericsson, M.; Huber, B.R.; Kurnick, S.A.; et al. Fatal Neurodissemination and SARS-CoV-2 Tropism in K18-hACE2 Mice Is Only Partially Dependent on hACE2 Expression. Viruses 2022, 14, 535. [Google Scholar] [CrossRef] [PubMed]
- Dinnon, K.H., 3rd; Leist, S.R.; Schäfer, A.; Edwards, C.E.; Martinez, D.R.; Montgomery, S.A.; West, A.; Yount, B.L., Jr.; Hou, Y.J.; Adams, L.E.; et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Kareinen, L.; Smura, T.; Freitag, T.L.; Jha, S.K.; Alitalo, K.; Meri, S.; Sironen, T.; Saksela, K.; Strandin, T.; et al. Common Laboratory Mice Are Susceptible to Infection with the SARS-CoV-2 Beta Variant. Viruses 2021, 13, 2263. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Yoon, C.; Hu, J.-C.; Wen, L.; Hu, B.; Yang, D.; Wang, Y.; Hou, Y.; et al. Emerging SARS-CoV-2 variants expand species tropism to murines. eBioMedicine 2021, 73, 103643. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 585, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Williamson, B.N.; Feldmann, F.; Schwarz, B.; Meade-White, K.; Porter, D.P.; Schulz, J.; van Doremalen, N.; Leighton, I.; Yinda, C.K.; Pérez-Pérez, L.; et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020, 585, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Munnink, B.B.O.; De Meulder, D.; Van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef]
- Cross, R.W.; Agans, K.N.; Prasad, A.N.; Borisevich, V.; Woolsey, C.; Deer, D.J.; Dobias, N.S.; Geisbert, J.B.; Fenton, K.A.; Geisbert, T.W. Intranasal exposure of African green monkeys to SARS-CoV-2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol. J. 2020, 17, 125. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.H.; Fung, K.; Choy, K.-T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef]
- Port, J.R.; Yinda, C.K.; Owusu, I.O.; Holbrook, M.; Fischer, R.; Bushmaker, T.; Avanzato, V.A.; Schulz, J.E.; van Doremalen, N.; Clancy, C.S.; et al. SARS-CoV-2 disease severity and transmission efficiency is increased for airborne compared to fomite exposure in Syrian hamsters. Nat. Commun. 2021, 12, 4985. [Google Scholar] [CrossRef]
- Yen, H.L.; Valkenburg, S.; Sia, S.F.; Choy, K.T.; Peiris, J.M.; Wong, K.H.; Crossland, N.; Douam, F.; Nicholls, J.M. Cellular tropism of SARS-CoV-2 in the respiratory tract of Syrian hamsters and B6.Cg-Tg(K18-ACE2)2Prlmn/J transgenic mice. Vet. Pathol. 2022, 59, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Haddock, E.; Feldmann, F.; Shupert, W.L.; Feldmann, H. Inactivation of SARS-CoV-2 Laboratory Specimens. Am. J. Trop. Med. Hyg. 2021, 104, 2195–2198. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Chamanza, R.; Wright, J.A. A Review of the Comparative Anatomy, Histology, Physiology and Pathology of the Nasal Cavity of Rats, Mice, Dogs and Non-human Primates. Relevance to Inhalation Toxicology and Human Health Risk Assessment. J. Comp. Pathol. 2015, 153, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.A.; Swenberg, J.A.; Fields, S.; Popp, J.A. Comparative morphometry of the nasal cavity in rats and mice. J. Anat. 1982, 135, 83–88. [Google Scholar] [PubMed]
- Harkema, J.R. Comparative aspects of nasal airway anatomy: Relevance to inhalation toxicology. Toxicol. Pathol. 1991, 19, 321–336. [Google Scholar] [CrossRef] [PubMed]






| Immunohistochemical Detection of SARS-CoV-2 N-Protein | |||
|---|---|---|---|
| Ciliated Respiratory Epithelium | Neurosensory Olfactory Epithelium | Lower Respiratory Tree | |
| 6 h Post Inoculation | − | − | − |
| 12 h Post Inoculation | Rare | − | Rare |
| 36 h Post Inoculation | + | − | + |
| 48 h Post Inoculation | + | + | + |
| 72 h Post Inoculation | Rare | + | + |
| Irradiated Virus Control | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clancy, C.S.; Meade-White, K.; Shaia, C.; Saturday, G.; Feldmann, H.; Rosenke, K. Histopathologic Characterization of Experimental Peracute SARS-CoV-2 Infection in the Syrian Hamster. Vet. Sci. 2023, 10, 536. https://doi.org/10.3390/vetsci10090536
Clancy CS, Meade-White K, Shaia C, Saturday G, Feldmann H, Rosenke K. Histopathologic Characterization of Experimental Peracute SARS-CoV-2 Infection in the Syrian Hamster. Veterinary Sciences. 2023; 10(9):536. https://doi.org/10.3390/vetsci10090536
Chicago/Turabian StyleClancy, Chad S., Kimberly Meade-White, Carl Shaia, Greg Saturday, Heinz Feldmann, and Kyle Rosenke. 2023. "Histopathologic Characterization of Experimental Peracute SARS-CoV-2 Infection in the Syrian Hamster" Veterinary Sciences 10, no. 9: 536. https://doi.org/10.3390/vetsci10090536