Trehalose Production Using Three Extracellular Enzymes Produced via One-Step Fermentation of an Engineered Bacillus subtilis Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Primers
2.2. Construction of the Engineered Strains
2.3. Fermentation Analysis of Recombinant B. subtilis Strains
2.4. Analysis of Protein Expression and Activity of the Recombinant Enzymes
2.5. Enzymatic Trehalose Production from Maltodextrin
2.6. Determination of the Activity of Mixed Enzymes Obtained via One-Step Fermentation
2.7. Detected Stability of the Strain’s Enzyme Production and Complex Enzymes
3. Results and Discussion
3.1. Screening of the Malto-Oligosyltrehalose Synthase and Malto-Oligosyltrehalose Trehalohydrolase from Different Sources
3.2. Neutral Pullulanase PulA Enhanced Trehalose Conversion Rate
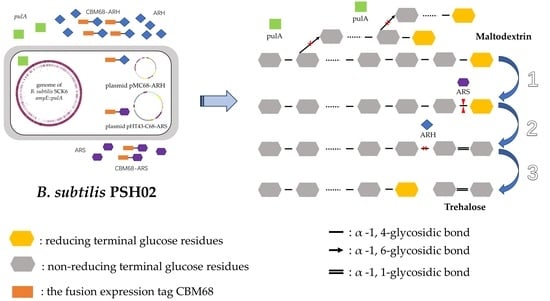
3.3. Construction of the Engineered Strains for Co-Production of the Three Enzymes
3.4. Properties of the Engineered Strain B. subtilis PSH02 for Trehalose Enzymatic Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qiao, Y.; Wang, W.; Lu, X. Engineering cyanobacteria as cell factories for direct trehalose production from CO2. Metab. Eng. 2020, 62, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chi, Z.; Liu, G.L.; Xue, S.J.; Wang, Z.P.; Hu, Z.; Chi, Z.M. Improved pullulan production by a mutant of Aureobasidium melanogenum TN3-1 from a natural honey and capsule shell preparation. Int. J. Biol. Macromol. 2019, 141, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Vanaporn, M.; Titball, R.W. Trehalose and bacterial virulence. Virulence 2020, 11, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Tapia, H.; Goddard, J.; Gibney, P. Trehalose and its applications in the food industry. Compr. Rev. Food Sci. Food Saf. 2022, 21, 5004–5037. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, X.; Liu, F.; Xie, J.; Zhu, Q.; Tan, S. Trehalose in Biomedical Cryopreservation-Properties, Mechanisms, Delivery Methods, Applications, Benefits, and Problems. ACS Biomater. Sci. Eng. 2023, 9, 1190–1204. [Google Scholar] [CrossRef]
- MacIntyre, A.M.; Barth, J.X.; Pellitteri Hahn, M.C.; Scarlett, C.O.; Genin, S.; Allen, C. Trehalose Synthesis Contributes to Osmotic Stress Tolerance and Virulence of the Bacterial Wilt Pathogen Ralstonia solanacearum. Mol. Plant-Microbe Interact. 2020, 33, 462–473. [Google Scholar] [CrossRef]
- Chen, A.; Gibney, P. Dietary Trehalose as a Bioactive Nutrient. Nutrients 2023, 15, 1393. [Google Scholar] [CrossRef]
- Sharma, E.; Shruti, P.S.; Singh, S.; Singh, T.; Kaur, P.; Jodha, B.; Srivastava, Y.; Munshi, A.; Singh, S. Trehalose and its diverse biological potential. Curr. Protein Pept. Sci. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Trakarnpaiboon, S.; Champreda, V. Integrated Whole-Cell Biocatalysis for Trehalose Production from Maltose Using Permeabilized Pseudomonas monteilii Cells and Bioremoval of Byproduct. J. Microbiol. Biotechnol. 2022, 32, 1054–1063. [Google Scholar] [CrossRef]
- Su, L.; Yao, K.; Wu, J. Improved Activity of Sulfolobus acidocaldarius Maltooligosyltrehalose Synthase through Directed Evolution. J. Agric. Food Chem. 2020, 68, 4456–4463. [Google Scholar] [CrossRef]
- Su, L.; Wu, S.; Feng, J. High-efficiency expression of Sulfolobus acidocaldarius maltooligosyl trehalose trehalohydrolase in Escherichia coli through host strain and induction strategy optimization. Bioprocess Biosyst. Eng. 2019, 42, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Su, L.; Xu, F.; Xia, Y. Improved Thermostability of Maltooligosyltrehalose Synthase from Arthrobacter ramosus by Directed Evolution and Site-Directed Mutagenesis. J. Agric. Food Chem. 2019, 67, 5587–5595. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Su, P.C.; Chen, P.T. Production and characterization of a recombinant thermophilic trehalose synthase from Thermus antranikianii. J. Biosci. Bioeng. 2020, 129, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Singh, S. A Novel Trehalose Synthase for the Production of Trehalose and Trehalulose. Microbiol. Spectr. 2021, 9, e0133321. [Google Scholar] [CrossRef]
- Zhu, L.; Shen, B.; Song, Z.; Jiang, L. Permeabilized TreS-Expressing Bacillus subtilis Cells Decorated with Glucose Isomerase and a Shell of ZIF-8 as a Reusable Biocatalyst for the Coproduction of Trehalose and Fructose. J. Agric. Food Chem. 2020, 68, 4464–4472. [Google Scholar] [CrossRef]
- Dong, L.; Lin, X.; Yu, D.; Huang, L.; Wang, B.; Pan, L. High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation. J. Ind. Microbiol. Biotechnol. 2019, 47, 133–144. [Google Scholar] [CrossRef]
- Liu, H.; Yang, S.; Wang, X.; Wang, T. Production of trehalose with trehalose synthase expressed and displayed on the surface of Bacillus subtilis spores. Microb. Cell Factories 2013, 18, 100. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Yang, S.; Wang, R.; Wang, T. Saturation mutagenesis and self-inducible expression of trehalose synthase in Bacillus subtilis. Biotechnol. Prog. 2019, 35, e2826. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Yang, S.; Wang, R.; Wang, T. Improved Expression and Optimization of Trehalose Synthase by Regulation of Pglv in Bacillus subtilis. Sci. Rep. 2019, 9, 6585. [Google Scholar] [CrossRef]
- Kobayashi, K.; Komeda, T.; Miura, Y.; Kettoku, M.; Kato, M. Production of Trehalose from Starch by Novel Trehalose-Producing Enzymes from Sulfolobus solfataricus KM1. J. Ferment. Bioeng. 1997, 83, 296–298. [Google Scholar] [CrossRef]
- Jiang, X.R.; Lin, Y.F.; Chen, P.T. Trehalose production via merged secretion, purification, and immobilization of trehalose synthase in Bacillus subtilis. Taiwan Inst. Chem. Eng. 2018, 82, 23–27. [Google Scholar] [CrossRef]
- Shu, W.; Zheng, H.; Fu, X.; Zhen, J.; Tan, M.; Xu, J.; Zhao, X.; Yang, S.; Song, H.; Ma, Y. Enhanced Heterologous Production of Glycosyltransferase UGT76G1 by Co-Expression of Endogenous prpD and malK in Escherichia coli and Its Transglycosylation Application in Production of Rebaudioside. Int. J. Mol. Sci. 2020, 21, 5752. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fu, X.; Zhao, X.; Xu, J.; Liu, Y.; Zheng, H.; Bai, W. Overexpression of a Thermostable α-Amylase through Genome Integration in Bacillus subtilis. Fermentation 2023, 9, 139. [Google Scholar] [CrossRef]
- Xu, J.; Ren, F.; Huang, C.H.; Zheng, Y.; Zhen, J.; Sun, H.; Ko, T.P.; He, M.; Chen, C.C.; Chan, H.C.; et al. Functional and structural studies of pullulanase from Anoxybacillus sp. LM18-11. Proteins 2014, 82, 1685–1693. [Google Scholar] [CrossRef]
- Zhen, J.; Zheng, H.; Zhao, X.; Fu, X.; Yang, S.; Xu, J.; Song, H.; Ma, Y. Regulate the hydrophobic motif to enhance the non-classical secretory expression of Pullulanase PulA in Bacillus subtilis. Int. J. Biol. Macromol. 2021, 193, 238–246. [Google Scholar] [CrossRef]
- Zeng, Y.; Xu, J.; Fu, X.; Tan, M.; Liu, F.; Zheng, H.; Song, H. Effects of different carbohydrate-binding modules on the enzymatic properties of pullulanase. Int. J. Biol. Macromol. 2019, 137, 973–981. [Google Scholar] [CrossRef]
- Han, C.; Su, L.; Hong, R.; Wu, S. A comparative study of maltooligosyltrehalose synthase from Sulfolobus acidocaldarius expressed in Pichia pastoris and Escherichia coli. Process Biochem. 2017, 60, 35–41. [Google Scholar] [CrossRef]
- Yamamoto, T.; Maruta, K.; Watanabe, H.; Yamashita, H.; Kubota, M.; Fukuda, S.; Kurimoto, M. Trehalose-producing Operon treYZ from Arthrobacter ramosus S34. Biosci. Biotechnol. Biochem. 2001, 65, 1419–1423. [Google Scholar] [CrossRef]
- Fang, T.; Tseng, W.C.; Pan, C.; Chun, Y.; Wang, M. Protein Engineering of Sulfolobus solfataricus Maltooligosyltrehalose Synthase To Alter Its Selectivity. J. Agric. Food Chem. 2007, 55, 5588–5594. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Yang, J.; Fu, X.; Zhao, X.; Zhen, J.; Song, H.; Xu, J.; Zheng, H.; Bai, W. Trehalose Production Using Three Extracellular Enzymes Produced via One-Step Fermentation of an Engineered Bacillus subtilis Strain. Bioengineering 2023, 10, 977. https://doi.org/10.3390/bioengineering10080977
Sun X, Yang J, Fu X, Zhao X, Zhen J, Song H, Xu J, Zheng H, Bai W. Trehalose Production Using Three Extracellular Enzymes Produced via One-Step Fermentation of an Engineered Bacillus subtilis Strain. Bioengineering. 2023; 10(8):977. https://doi.org/10.3390/bioengineering10080977
Chicago/Turabian StyleSun, Xi, Jun Yang, Xiaoping Fu, Xingya Zhao, Jie Zhen, Hui Song, Jianyong Xu, Hongchen Zheng, and Wenqin Bai. 2023. "Trehalose Production Using Three Extracellular Enzymes Produced via One-Step Fermentation of an Engineered Bacillus subtilis Strain" Bioengineering 10, no. 8: 977. https://doi.org/10.3390/bioengineering10080977