An Efficient Approach to Predict Eye Diseases from Symptoms Using Machine Learning and Ranker-Based Feature Selection Methods
Abstract
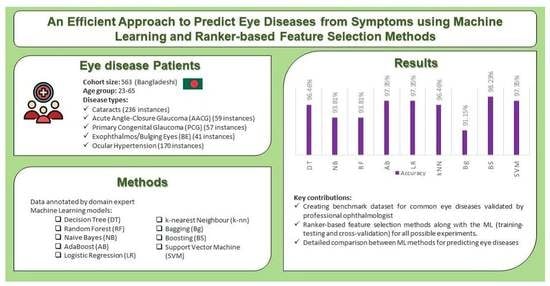
:1. Introduction
- Creating a benchmark dataset in the domain of eye diseases validated by professional ophthalmologists, that cam be applied to test ML, AI and Symptomatic analyses.
- Utilizing ranker-based feature selection methods to identify highly ranked symptoms among the five diseases.
- Experimenting with scenarios both with and without splitting the dataset and with several feature selection methods for better predictions.
- Compare the performance of classic ML methods to efficiently predict the occurrence of eye diseases.
2. Overview of Five Eye Diseases
2.1. Cataracts
2.2. Acute Angle-Closure Glaucoma (AACG)
2.3. Primary Congenital Glaucoma (PCG)
2.4. Exophthalmos or Bulging Eyes
2.5. Ocular Hypertension
3. Related Works
4. Research Methodology
4.1. Data Collection
4.2. Data Annotation
4.3. Feature Selection Methods
4.4. Data Splitting Strategies
4.5. Machine Learning Methods
5. Performance Measurement Indices
6. Experimental Results
6.1. Applying Feature Selection Methods
6.2. Experiments on Data Splitting and FS Methods
6.2.1. Experiment-1: Splitting + FS Applied
6.2.2. Experiment-2: Splitting + No FS Applied
6.2.3. Experiment-3: Cross-Validation + FS Applied
6.2.4. Experiment-4: Cross-Validation + No FS Applied
7. Discussion
7.1. Finding Significant Features
7.2. Finding Common Features
7.3. Comparison with Existing Works
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sutradhar, I.; Gayen, P.; Hasan, M.; Gupta, R.D.; Roy, T.; Sarker, M. Eye diseases: The neglected health condition among urban slum population of Dhaka, Bangladesh. BMC Ophthalmol. 2019, 19, 38. [Google Scholar] [CrossRef] [Green Version]
- Ayodele, T.O. Types of machine learning algorithms. New Adv. Mach. Learn. 2010, 3, 19–48. [Google Scholar]
- Mair, C.; Kadoda, G.; Lefley, M.; Phalp, K.; Schofield, C.; Shepperd, M.; Webster, S. An investigation of machine learning based prediction systems. J. Syst. Softw. 2000, 53, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, A. The production of prediction: What does machine learning want? Eur. J. Cult. Stud. 2015, 18, 429–445. [Google Scholar] [CrossRef] [Green Version]
- Hodge, W.G.; Whitcher, J.P.; Satariano, W. Risk factors for age-related cataracts. Epidemiol. Rev. 1995, 17, 336–346. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Wilkins, M.; Kim, T.; Malyugin, B.; Mehta, J.S. Cataracts. The Lancet 2017, 390, 600–612. [Google Scholar] [CrossRef]
- Petsas, A.; Chapman, G.; Stewart, R. Acute angle closure glaucoma—A potential blind spot in critical care. J. Intensive Care Soc. 2017, 18, 244–246. [Google Scholar] [CrossRef] [Green Version]
- Ko, F.; Papadopoulos, M.; Khaw, P.T. Primary congenital glaucoma. Prog. Brain Res. 2015, 221, 177–189. [Google Scholar] [PubMed]
- Badawi, A.H.; Al-Muhaylib, A.A.; Al Owaifeer, A.M.; Al-Essa, R.S.; Al-Shahwan, S.A. Primary congenital glaucoma: An updated review. Saudi J. Ophthalmol. 2019, 33, 382–388. [Google Scholar] [CrossRef]
- Moro, A.; Munhoz, R.P.; Arruda, W.O.; Raskin, S.; Teive, H.A.G. Clinical relevance of “bulging eyes” for the differential diagnosis of spinocerebellar ataxias. Arquivos de Neuro-Psiquiatria 2013, 71, 428–430. [Google Scholar] [CrossRef] [Green Version]
- Argus, W.A. Ocular hypertension and central corneal thickness. Ophthalmology 1995, 102, 1810–1812. [Google Scholar] [CrossRef] [PubMed]
- Muhit, M.; Karim, T.; Jahan, I.; Al Imam, M.H.; Das, M.C.; Khandaker, G. Epidemiology of eye diseases among children with disability in rural Bangladesh: A population-based cohort study. Dev. Med. Child Neurol. 2022, 64, 209–219. [Google Scholar] [CrossRef]
- Kadir, S.M.U.; Ali, M.; Islam, M.S.; Parvin, R.; Quadir, A.S.M.; Raihani, M.J.; Islam, A.R.; Ahmmed, S. Prevalence of Refractive Errors among Primary School Children in the Southern Region of Bangladesh. Community Based Med. J. 2022, 11, 41–45. [Google Scholar] [CrossRef]
- Sarki, R.; Ahmed, K.; Wang, H.; Zhang, Y.; Ma, J.; Wang, K. Image preprocessing in classification and identification of diabetic eye diseases. Data Sci. Eng. 2021, 6, 455–471. [Google Scholar] [CrossRef]
- Umesh, L.; Mrunalini, M.; Shinde, S. Review of image processing and machine learning techniques for eye disease detection and classification. Int. Res. J. Eng. Technol. 2016, 3, 547–551. [Google Scholar]
- Oda, M.; Yamaguchi, T.; Fukuoka, H.; Ueno, Y.; Mori, K. Automated eye disease classification method from anterior eye image using anatomical structure focused image classification technique. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 16–19 February 2020; Volume 11314, pp. 991–996. [Google Scholar]
- Fourcade, A.; Khonsari, R.H. Deep learning in medical image analysis: A third eye for doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef]
- Acharya, U.R.; Kannathal, N.; Ng, E.Y.K.; Min, L.C.; Suri, J.S. Computer-based classification of eye diseases. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006; pp. 6121–6124. [Google Scholar]
- Nazir, T.; Nawaz, M.; Rashid, J.; Mahum, R.; Masood, M.; Mehmood, A.; Hussain, A. Detection of diabetic eye disease from retinal images using a deep learning based CenterNet model. Sensors 2021, 21, 5283. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Shaik, N.S.; Naralasetti, V. Deep convolution feature aggregation: An application to diabetic retinopathy severity level prediction. Signal Image Video Process. 2021, 15, 923–930. [Google Scholar] [CrossRef]
- Khan, M.S.M.; Ahmed, M.; Rasel, R.Z.; Khan, M.M. Cataract detection using convolutional neural network with VGG-19 model. In Proceedings of the 2021 IEEE World AI IoT Congress (AIIoT), Seattle, WA, USA, 10–13 May 2021; pp. 0209–0212. [Google Scholar]
- Sarki, R.; Ahmed, K.; Wang, H.; Zhang, Y.; Wang, K. Convolutional neural network for multi-class classification of diabetic eye disease. EAI Endorsed Trans. Scalable Inf. Syst. 2022, 9, e15. [Google Scholar] [CrossRef]
- Pahuja, R.; Sisodia, U.; Tiwari, A.; Sharma, S.; Nagrath, P. A Dynamic Approach of Eye Disease Classification Using Deep Learning and Machine Learning Model. In Proceedings of Data Analytics and Management; Springer: Singapore, 2022; pp. 719–736. [Google Scholar]
- Malik, S.; Kanwal, N.; Asghar, M.N.; Sadiq, M.A.A.; Karamat, I.; Fleury, M. Data driven approach for eye disease classification with machine learning. Appl. Sci. 2019, 9, 2789. [Google Scholar] [CrossRef] [Green Version]
- Bitto, A.K.; Mahmud, I. Multi categorical of common eye disease detect using convolutional neural network: A transfer learning approach. Bull. Electr. Eng. Inform. 2022, 11, 2378–2387. [Google Scholar] [CrossRef]
- Verma, S.; Singh, L.; Chaudhry, M. Classifying red and healthy eyes using deep learning. Illumination 2019, 10, 525–531. [Google Scholar] [CrossRef]
- Hameed, S.; Ahmed, H.M. Eye diseases classification using back propagation with parabola learning rate. Al-Qadisiyah J. Pure Sci. 2021, 26, 1–9. [Google Scholar] [CrossRef]
- Bhadra, A.A.; Jain, M.; Shidnal, S.S. Automated detection of eye diseases. In Proceedings of the 2016 International Conference on Wireless Communications, Signal Processing and Networking (WiSPNET), Chennai, India, 23–25 March 2016; pp. 1341–1345. [Google Scholar] [CrossRef]
- Prasad, K.; Sajith, P.S.; Neema, M.; Madhu, L.; Priya, P.N. Multiple eye disease detection using Deep Neural Network. In Proceedings of the TENCON 2019—2019 IEEE Region 10 Conference (TENCON), Kochi, India, 17–20 October 2019; pp. 2148–2153. [Google Scholar] [CrossRef]
- Pearson, K. On the Criterion That a Given System of Deviations From the Probable in the Case of a Correlated System of Variables is Such That It Can Be Reasonably Supposed to Have Arisen From Random Sampling. Philos. Mag. 1900, 5, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Forman, G. An extensive empirical study of feature selection metrics for text classification. J. Mach. Learn. Res. 2003, 3, 1289–1305. [Google Scholar]
- Gao, Z.; Xu, Y.; Meng, F.; Qi, F.; Lin, L. Improved information gain-based feature selection for text categorization. In Proceedings of the 2014 4th International Conference on Wireless Communications, Vehicular Technology, Information Theory and Aerospace & Electronic Systems (VITAE), Aalborg, Denmark, 11–14 May 2014; pp. 11–14. [Google Scholar]
- Yu, L.; Liu, H. Feature selection for high-dimensional data: A fast correlation-based filter solution. In Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 856–863. [Google Scholar]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://en.wikipedia.org/wiki/Principal_component_analysis (accessed on 18 December 2022).
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Song, F.; Guo, Z.; Mei, D. Feature selection using principal component analysis. In Proceedings of the 2010 International Conference on System Science, Engineering Design and Manufacturing Informatization, Yichang, China, 12–14 November 2010; pp. 27–30. [Google Scholar]
- Kira, K.; Rendell, L.A. The feature selection problem: Traditional methods and a new algorithm. AAAI 1992, 2, 129–134. [Google Scholar]
- Abraham, M.T.; Satyam, N.; Lokesh, R.; Pradhan, B.; Alamri, A. Factors affecting landslide susceptibility mapping: Assessing the influence of different machine learning approaches, sampling strategies and data splitting. Land 2021, 10, 989. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Rish, I. An empirical study of the Naive Bayes classifier. In Proceedings of the JCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, DC, USA, 4–6 August 2001; Volume3, pp. 41–46. [Google Scholar]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient kNN classification with different numbers of nearest neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Myles, A.J.; Feudale, R.N.; Liu, Y.; Woody, N.A.; Brown, S.D. An introduction to decision tree modeling. J. Chemom. J. Chemom. Soc. 2004, 18, 275–285. [Google Scholar] [CrossRef]
- LaValley, M.P. Logistic regression. Circulation 2008, 117, 2395–2399. [Google Scholar] [CrossRef]
- Marouf, A.A.; Hasan, M.K.; Mahmud, H. Comparative analysis of feature selection algorithms for computational personality prediction from social media. IEEE Trans. Comput. Soc. Syst. 2020, 7, 587–599. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Jonkman, M.; Karim, A.; Shamrat, F.J.M.; Ignatious, E.; Shultana, S.; Beeravolu, A.R.; De Boer, F. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 2021, 9, 19304–19326. [Google Scholar] [CrossRef]






| Properties | Amount/Values |
|---|---|
| No. of patients | 563 |
| Age group of patients | 23–65 |
| Gender of patients | Male or Female |
| No. of instances in the dataset | 563 |
| Data collection process | In-person interview with patients |
| Type of interview | Semi-structured interviews |
| Type of pre-defined questionnaire | Binary closed questions (Yes/No) |
| Types of eye diseases |
|
| No. | Attributes | Properties |
|---|---|---|
| a1 | Cloudy, blurry or foggy vision | The values are either 0 or 1 |
| a2 | Pressure in eye | |
| a3 | Injury to the eye | |
| a4 | Excessive dryness | |
| a5 | Red eye | |
| a6 | Cornea increased in size | |
| a7 | Problem in identifying color | |
| a8 | Double vision | |
| a9 | Myopia | |
| a10 | Trouble with glasses | |
| a11 | Hard to see in the dark | |
| a12 | Visible whiteness | |
| a13 | Mass pain | |
| a14 | Vomiting | |
| a15 | Water drops from eyes continuously | |
| a16 | Presence of light when eye lid closes | |
| a17 | Family history of similar disease | |
| a18 | Age +40 | Biomarker (0 or 1) |
| a19 | Diabetes |
| Ranking Score | Attributes (Attribute Number) |
|---|---|
| 0.6218 | Cloudy, blurry or foggy vision (a1) |
| 0.5583 | Problem in identifying color (a7) |
| 0.5340 | Double vision (a8) |
| 0.5148 | Water drops from eyes continuously (a15) |
| 0.5114 | Pressure in eye (a2) |
| 0.5073 | Hard to see in the dark (a11) |
| 0.4991 | Myopia (a9) |
| 0.3052 | Injury to the eye (a3) |
| 0.2403 | Mass pain (a13), |
| 0.2279 | Red eye (a5) |
| 0.2258 | Vomiting (a14) |
| 0.2225 | Cornea increased in size (a6) |
| 0.2225 | Presents of light when eyelid close (a16) |
| 0.2061 | Visible whiteness (a12) |
| 0.2023 | Excessive dryness (a4) |
| 0.1539 | 40+ Age (a18) |
| 0.1531 | Family history of similar disease (a17) |
| 0.1525 | Diabetes (a19) |
| 0.0524 | Trouble with glasses (a10) |
| Ranking Score | Attributes (Attribute Number) |
|---|---|
| 0.8000 | Cloudy, blurry or foggy vision (a1) |
| 0.6182 | Problem in identifying color (a7) |
| 0.5587 | Water drops from eyes continuously (a15) |
| 0.5434 | Double vision (a8) |
| 0.5430 | Pressure in eye (a2) |
| 0.5216 | Myopia (a9) |
| 0.4848 | Hard to see in the dark (a11) |
| 0.3477 | Injury to the eye (a3) |
| 0.2326 | Mass pain (a13) |
| 0.2293 | Visible whiteness (a12) |
| 0.2196 | Excessive dryness (a4) |
| 0.2130 | Cornea increased in size (a6) |
| 0.2130 | Presents of light when eyelid close (a16) |
| 0.2108 | Red eye (a5) |
| 0.2004 | Vomiting (a14) |
| 0.0749 | 40+ Age (a18) |
| 0.0730 | Family history of similar disease (a17) |
| 0.0718 | Diabetes (a19) |
| 0.0000 | Trouble with glasses (a10) |
| Ranking Score | Attributes (Attribute Number) |
|---|---|
| 0.6194 | Cloudy, blurry or foggy vision (a1) |
| 0.4196 | Pressure in eye (a2) |
| 0.4181 | Problem in identifying color (a7) |
| 0.3682 | Injury to the eye (a3) |
| 0.3383 | Myopia (a9) |
| 0.3253 | Double vision (a8) |
| 0.3186 | Water drops from eyes continuously (a15) |
| 0.3137 | Hard to see in the dark (a11) |
| 0.1129 | 40+ Age (a18) |
| 0.1032 | Visible whiteness (a12) |
| 0.1024 | Red eye (a5) |
| 0.1013 | Cornea increased in size (a6) |
| 0.0952 | Diabetes (a19) |
| 0.0915 | Mass pain (a13) |
| 0.0875 | Excessive dryness (a4) |
| 0.0826 | Vomiting (a14) |
| 0.0773 | Presents of light when eyelid close (a16) |
| 0.0616 | Family history of similar disease (a17) |
| 0.0218 | Trouble with glasses (a10) |
| Ranking Score | Attributes (Attribute Number) |
|---|---|
| 0.7619 | Cloudy, blurry or foggy vision (a1) |
| 0.5981 | Injury to the eye (a3) |
| 0.5054 | Excessive dryness (a4) |
| 0.4171 | Presents of light when eyelid close (a16) |
| 0.365 | Trouble with glasses (a10) |
| 0.3175 | Pressure in eye (a2) |
| 0.2719 | 40+ Age (a18) |
| 0.2306 | Family history of similar disease (a17) |
| 0.1976 | Vomiting (a14) |
| 0.1678 | Mass pain (a13) |
| 0.1389 | Red eye (a5) |
| 0.1136 | Double vision (a8) |
| 0.09 | Cornea increased in size (a6) |
| 0.0719 | Problem in identifying color (a7) |
| 0.0549 | Myopia (a9) |
| 0.0388 | Hard to see in the dark (a11) |
| Model | 66–34% Split | 75–25% Split | 80–20% Split | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | PR | SEN | F1-Score | ACC | PR | SEN | F1-Score | ACC | PR | SEN | F1-Score | ACC |
| DT | 0.97 | 0.97 | 0.97 | 96.875% | 0.97 | 0.97 | 0.97 | 97.163% | 0.97 | 0.96 | 0.96 | 96.460% |
| NB | 0.95 | 0.94 | 0.94 | 94.271% | 0.96 | 0.95 | 0.95 | 95.035% | 0.95 | 0.94 | 0.94 | 93.805% |
| RF | 1.00 | 0.81 | 0.88 | 81.25% | 1.00 | 0.82 | 0.89 | 81.56% | 1.00 | 0.78 | 0.87 | 77.876% |
| AB | 0.98 | 0.98 | 0.98 | 97.51% | 0.98 | 0.98 | 0.98 | 98.05% | 0.98 | 0.98 | 0.98 | 97.69% |
| LR | 0.98 | 0.98 | 0.98 | 97.917% | 0.99 | 0.99 | 0.99 | 98.582% | 0.97 | 0.97 | 0.97 | 97.345% |
| k-NN | 0.97 | 0.97 | 0.97 | 96.875% | 0.97 | 0.97 | 0.97 | 97.163% | 0.96 | 0.96 | 0.96 | 96.46% |
| Bagging | 0.91 | 0.91 | 0.91 | 91.513% | 0.91 | 0.84 | 0.82 | 91.56% | 0.80 | 0.90 | 0.89 | 90.088% |
| XGBoost | 0.98 | 0.98 | 0.98 | 98.579% | 0.99 | 0.99 | 0.99 | 98.581% | 0.98 | 0.98 | 0.98 | 98.23% |
| SVM | 0.64 | 0.74 | 0.65 | 74.479% | 0.63 | 0.74 | 0.64 | 73.759% | 0.59 | 0.70 | 0.59 | 69.912% |
| Cross Fold Validation with FS Method | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methods | Feature Selection Methods | 3-fold | 5-fold | 10-fold | |||||||||
| P | R | F1 | ACC | P | R | F1 | ACC | P | R | F1 | ACC | ||
| DT | PC | 0.914 | 0.892 | 0.892 | 89.17% | 0.918 | 0.899 | 0.900 | 89.88% | 0.913 | 0.897 | 0.896 | 89.70% |
| IG | 0.902 | 0.893 | 0.896 | 89.34% | 0.906 | 0.897 | 0.899 | 89.70% | 0.907 | 0.899 | 0.899 | 89.88% | |
| PCA | 0.925 | 0.911 | 0.912 | 91.12% | 0.926 | 0.913 | 0.914 | 91.30% | 0.926 | 0.913 | 0.914 | 91.30% | |
| Relief | 0.916 | 0.899 | 0.899 | 89.88% | 0.921 | 0.899 | 0.899 | 89.88% | 0.918 | 0.899 | 0.899 | 89.88% | |
| All * | 0.960 | 0.950 | 0.950 | 94.85% | 0.960 | 0.960 | 0.960 | 96.81% | 0.930 | 0.930 | 0.920 | 96.98% | |
| NB | PC | 0.920 | 0.909 | 0.907 | 90.94% | 0.923 | 0.909 | 0.907 | 90.94% | 0.922 | 0.909 | 0.907 | 90.94% |
| IG | 0.924 | 0.915 | 0.913 | 91.47% | 0.910 | 0.909 | 0.909 | 90.94% | 0.898 | 0.899 | 0.898 | 89.88% | |
| PCA | 0.924 | 0.911 | 0.913 | 91.12% | 0.925 | 0.911 | 0.913 | 91.12% | 0.926 | 0.911 | 0.913 | 91.12% | |
| Relief | 0.916 | 0.904 | 0.902 | 90.41% | 0.912 | 0.901 | 0.899 | 90.05% | 0.916 | 0.902 | 0.901 | 90.23% | |
| All * | 0.960 | 0.960 | 0.960 | 95.56% | 0.960 | 0.960 | 0.950 | 95.92% | 0.950 | 0.930 | 0.390 | 95.74% | |
| RF | PC | 0.920 | 0.906 | 0.905 | 90.59% | 0.923 | 0.911 | 0.910 | 91.12% | 0.919 | 0.908 | 0.907 | 90.76% |
| IG | 0.908 | 0.902 | 0.901 | 90.23% | 0.923 | 0.915 | 0.915 | 91.47% | 0.918 | 0.909 | 0.909 | 90.94% | |
| PCA | 0.911 | 0.899 | 0.901 | 89.88% | 0.895 | 0.890 | 0.890 | 88.99% | 0.899 | 0.892 | 0.893 | 89.17% | |
| Relief | 0.923 | 0.904 | 0.902 | 90.41% | 0.924 | 0.904 | 0.904 | 90.41% | 0.923 | 0.904 | 0.903 | 90.41% | |
| All * | 0.990 | 0.990 | 0.990 | 98.40% | 1.000 | 1.000 | 1.000 | 98.58% | 1.000 | 1.000 | 1.000 | 97.87% | |
| AB | PC | 0.913 | 0.901 | 0.900 | 90.05% | 0.923 | 0.908 | 0.908 | 90.80% | 0.921 | 0.908 | 0.907 | 90.76% |
| IG | 0.911 | 0.902 | 0.901 | 90.23% | 0.913 | 0.904 | 0.905 | 90.41% | 0.925 | 0.913 | 0.913 | 91.30% | |
| PCA | 0.895 | 0.890 | 0.890 | 88.99% | 0.909 | 0.901 | 0.901 | 90.05% | 0.905 | 0.897 | 0.898 | 89.70% | |
| Relief | 0.920 | 0.902 | 0.901 | 90.23% | 0.926 | 0.909 | 0.908 | 90.94% | 0.920 | 0.904 | 0.903 | 90.41% | |
| All * | 0.975 | 0.975 | 0.975 | 97.51% | 0.980 | 0.980 | 0.980 | 98.05% | 0.977 | 0.977 | 0.977 | 97.69% | |
| LR | PC | 0.919 | 0.902 | 0.904 | 90.23% | 0.924 | 0.911 | 0.911 | 91.12% | 0.922 | 0.909 | 0.909 | 90.94% |
| IG | 0.921 | 0.909 | 0.910 | 90.94% | 0.913 | 0.908 | 0.908 | 90.76% | 0.927 | 0.917 | 0.917 | 91.65% | |
| PCA | 0.900 | 0.893 | 0.895 | 89.34% | 0.904 | 0.899 | 0.900 | 89.88% | 0.911 | 0.902 | 0.904 | 90.23% | |
| Relief | 0.927 | 0.911 | 0.912 | 91.12% | 0.926 | 0.909 | 0.909 | 90.94% | 0.930 | 0.913 | 0.913 | 91.30% | |
| All * | 0.990 | 0.980 | 0.950 | 98.58% | 1.000 | 1.000 | 1.000 | 98.94% | 1.000 | 1.000 | 1.000 | 98.94% | |
| k-NN | PC | 0.930 | 0.915 | 0.913 | 91.47% | 0.929 | 0.917 | 0.915 | 91.65% | 0.926 | 0.915 | 0.912 | 91.47% |
| IG | 0.908 | 0.908 | 0.907 | 90.76% | 0.912 | 0.906 | 0.907 | 90.59% | 0.926 | 0.915 | 0.915 | 91.47% | |
| PCA | 0.892 | 0.886 | 0.887 | 88.63% | 0.906 | 0.899 | 0.899 | 89.88% | 0.901 | 0.895 | 0.896 | 89.52% | |
| Relief | 0.929 | 0.908 | 0.908 | 90.76% | 0.925 | 0.906 | 0.906 | 90.59% | 0.922 | 0.902 | 0.902 | 90.23% | |
| All * | 0.960 | 0.960 | 0.950 | 96.45% | 0.980 | 0.970 | 0.970 | 96.27% | 1.000 | 1.000 | 1.000 | 96.63% | |
| Bagging | PC | 0.914 | 0.897 | 0.896 | 89.70% | 0.926 | 0.909 | 0.907 | 90.94% | 0.925 | 0.911 | 0.910 | 91.00% |
| IG | 0.909 | 0.892 | 0.889 | 89.17% | 0.898 | 0.897 | 0.894 | 89.70% | 0.897 | 0.895 | 0.893 | 89.30% | |
| PCA | 0.911 | 0.899 | 0.899 | 89.88% | 0.909 | 0.895 | 0.895 | 89.52% | 0.905 | 0.890 | 0.889 | 88.99% | |
| Relief | 0.905 | 0.885 | 0.885 | 88.45% | 0.911 | 0.897 | 0.893 | 89.70% | 0.914 | 0.897 | 0.896 | 89.70% | |
| All * | 0.930 | 0.940 | 0.970 | 95.06% | 0.940 | 0.960 | 0.950 | 95.58% | 0.940 | 0.950 | 0.890 | 95.58% | |
| XGBoost | PC | 0.920 | 0.904 | 0.905 | 90.41% | 0.926 | 0.911 | 0.912 | 91.12% | 0.926 | 0.911 | 0.911 | 91.12% |
| IG | 0.929 | 0.917 | 0.917 | 91.65% | 0.920 | 0.911 | 0.913 | 91.12% | 0.924 | 0.913 | 0.915 | 91.30% | |
| PCA | 0.916 | 0.906 | 0.907 | 90.59% | 0.915 | 0.906 | 0.907 | 90.59% | 0.918 | 0.908 | 0.909 | 90.76% | |
| Relief | 0.929 | 0.913 | 0.913 | 91.30% | 0.923 | 0.908 | 0.908 | 90.76% | 0.927 | 0.909 | 0.910 | 90.94% | |
| All * | 0.980 | 0.980 | 0.980 | 98.58% | 0.990 | 0.990 | 0.990 | 98.58% | 0.980 | 0.980 | 0.980 | 98.05% | |
| SVM | PC | 0.934 | 0.911 | 0.907 | 91.12% | 0.935 | 0.913 | 0.907 | 91.30% | 0.928 | 0.911 | 0.905 | 91.12% |
| IG | 0.917 | 0.909 | 0.911 | 90.94% | 0.913 | 0.906 | 0.907 | 90.59% | 0.898 | 0.890 | 0.891 | 88.99% | |
| PCA | 0.904 | 0.888 | 0.888 | 88.81% | 0.902 | 0.890 | 0.888 | 88.99% | 0.899 | 0.892 | 0.891 | 89.17% | |
| Relief | 0.935 | 0.911 | 0.909 | 91.12% | 0.932 | 0.908 | 0.906 | 90.76% | 0.931 | 0.908 | 0.906 | 90.76% | |
| All * | 0.980 | 0.980 | 0.980 | 98.76% | 1.000 | 1.000 | 1.000 | 98.94% | 1.000 | 1.000 | 1.000 | 99.11% | |
| Model | 3-fold | 5-fold | 10-fold | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | PR | SEN | F1-Score | ACC | PR | SEN | F1-Score | ACC | PR | SEN | F1-Score | ACC |
| DT | 0.96 | 0.95 | 0.95 | 94.850% | 0.96 | 0.96 | 0.96 | 96.805% | 0.93 | 0.93 | 0.92 | 96.980% |
| NB | 0.96 | 0.96 | 0.96 | 95.561% | 0.96 | 0.96 | 0.95 | 95.915% | 0.95 | 0.93 | 0.93 | 95.742% |
| RF | 0.99 | 0.99 | 0.99 | 98.402% | 1.00 | 1.00 | 1.00 | 98.581% | 1.00 | 1.00 | 1.00 | 97.870% |
| AB | 0.98 | 0.98 | 0.98 | 97.510% | 0.98 | 0.98 | 0.98 | 98.050% | 0.98 | 0.98 | 0.98 | 97.690% |
| LR | 0.99 | 0.98 | 0.95 | 98.579% | 1.00 | 1.00 | 1.00 | 98.936% | 1.00 | 1.00 | 1.00 | 98.938% |
| k-NN | 0.96 | 0.96 | 0.95 | 96.447% | 0.98 | 0.97 | 0.97 | 96.271% | 1.00 | 1.00 | 1.00 | 96.626% |
| Bagging | 0.93 | 0.94 | 0.97 | 95.062% | 0.94 | 0.96 | 0.95 | 95.579% | 0.94 | 0.95 | 0.89 | 95.578% |
| XGBoost | 0.98 | 0.98 | 0.98 | 98.579% | 0.99 | 0.99 | 0.99 | 98.581% | 0.98 | 0.98 | 0.98 | 98.051% |
| SVM | 0.98 | 0.98 | 0.98 | 98.756% | 1.00 | 1.00 | 1.00 | 98.936% | 1.00 | 1.00 | 1.00 | 99.110% |
| Existing Works | Features Used | Feature Selection Used | Methods | Evaluation |
|---|---|---|---|---|
| Nazir et al. [19] | Extracted features using Densenet-100 | No | Improved CenterNet | Accuracy (using Aptos-2019 dataset: 97.93%, using IDrID dataset: 98.10%) |
| Bodapati et al. [20] | Feature fusion | No | Deep neural Network | Accuracy (using Aptos-2019 dataset: 84.31%) |
| Khan et al. [21] | Manual extracted retinal features | No | CNN with VGG-19 | Accuracy (97.47%) |
| Sarki et al. [22] | None | No | CNN with RMSprop Optmizer | Accuracy (81.33%) |
| Pahuja et al. [23] | None | No | SVM and CNN | Accuracy (SVM:87.5% and CNN: 85.42%) |
| Malik et al. [24] | None | No | DT, NB, RF and NN | Accuracy (RF: 86.63%) |
| Proposed | None | Yes (PC, IG, PCA, Relief) | DT, NB, RF, AB, LR, k-NN, Bagging, XGBoost and SVM | Accuracy (Highest Accuracy Obtained: 99.11% (SVM)) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marouf, A.A.; Mottalib, M.M.; Alhajj, R.; Rokne, J.; Jafarullah, O. An Efficient Approach to Predict Eye Diseases from Symptoms Using Machine Learning and Ranker-Based Feature Selection Methods. Bioengineering 2023, 10, 25. https://doi.org/10.3390/bioengineering10010025
Marouf AA, Mottalib MM, Alhajj R, Rokne J, Jafarullah O. An Efficient Approach to Predict Eye Diseases from Symptoms Using Machine Learning and Ranker-Based Feature Selection Methods. Bioengineering. 2023; 10(1):25. https://doi.org/10.3390/bioengineering10010025
Chicago/Turabian StyleMarouf, Ahmed Al, Md Mozaharul Mottalib, Reda Alhajj, Jon Rokne, and Omar Jafarullah. 2023. "An Efficient Approach to Predict Eye Diseases from Symptoms Using Machine Learning and Ranker-Based Feature Selection Methods" Bioengineering 10, no. 1: 25. https://doi.org/10.3390/bioengineering10010025