Identification and Antimicrobial Activity of Medium-Sized and Short Peptides from Yellowfin Tuna (Thunnus albacares) Simulated Gastrointestinal Digestion
Abstract
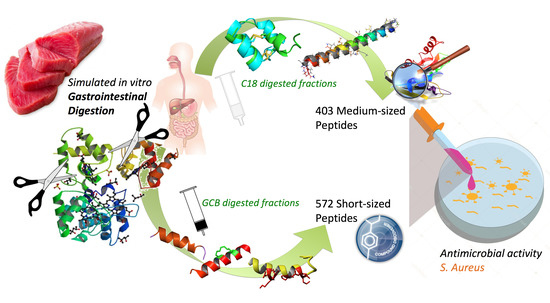
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Standards
2.2. Fish Protein Extraction
2.3. Simulated In Vitro Gastrointestinal Digestion
2.4. Hydrolyzed Peptide Purification
2.4.1. Medium-Sized Peptides Purification
2.4.2. Short-Sized Peptide Purification
2.5. Chromatography–Mass Spectrometry Analysis of Peptide Samples
2.5.1. Analysis of Medium-Sized Peptides by NanoHPLC–MS/MS
2.5.2. Analysis of Short-Sized Peptides by UHPLC–MS/MS
2.6. Qualitative Antimicrobial Assay
2.7. Quantitative Antimicrobial Assay—Minimum Inhibiting Concentration (MIC) Determination
2.8. Peptide Identification
2.8.1. Medium-Sized Peptide Identification
2.8.2. Short Peptide Identification
3. Results and Discussion
3.1. Antimicrobial Activity of C18 and GCB Digested Fractions
3.2. Characterization of Hydrolyzed Medium-Sized Peptides in Fish
3.3. Characterization of Hydrolyzed Short-Sized Peptides in Fish
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yathisha, U.G.; Bhat, I.; Karunasagar, I.; Mamatha, B.S. Antihypertensive activity of fish protein hydrolysates and its peptides. Crit. Rev. Food Sci. Nutr. 2019, 59, 2363–2374. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A.S. A review of fish-derived antioxidant and antimicrobial peptides: Their production, assessment, and applications. Peptides 2012, 33, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.; Ng, T.; Wong, J. Marine Peptides: Bioactivities and Applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Phelan, M.; Aherne, A.; FitzGerald, R.J.; O’Brien, N.M. Casein-derived bioactive peptides: Biological effects, industrial uses, safety aspects and regulatory status. Int. Dairy J. 2009, 19, 643–654. [Google Scholar] [CrossRef]
- Kang, H.K.; Lee, H.H.; Seo, C.H.; Park, Y. Antimicrobial and Immunomodulatory Properties and Applications of Marine-Derived Proteins and Peptides. Mar. Drugs 2019, 17, 350. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, P.; Vanhoye, D.; Amiche, M. Molecular strategies in biological evolution of antimicrobial peptides. Peptides 2003, 24, 1669–1680. [Google Scholar] [CrossRef]
- Patel, S.; Akhtar, N. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed. Pharmacother. 2017, 95, 1276–1283. [Google Scholar] [CrossRef]
- Valero, Y.; Saraiva-Fraga, M.; Costas, B.; Guardiola, F.A. Antimicrobial peptides from fish: Beyond the fight against pathogens. Rev. Aquac. 2020, 12, 224–253. [Google Scholar] [CrossRef]
- Aoki, W.; Ueda, M. Characterization of Antimicrobial Peptides toward the Development of Novel Antibiotics. Pharmaceuticals 2013, 6, 1055–1081. [Google Scholar] [CrossRef] [Green Version]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masso-Silva, J.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, D.; Jena, R.; Choudhury, P.K.; Pattnaik, R.; Mohapatra, S.; Saini, M.R. Milk Derived Antimicrobial Bioactive Peptides: A Review. Int. J. Food Prop. 2016, 19, 837–846. [Google Scholar] [CrossRef]
- Meisel, H.; FitzGerald, R.J. Biofunctional Peptides from Milk Proteins: Mineral Binding and Cytomodulatory Effects. Curr. Pharm. Des. 2003, 9, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Ejiri, M.; Mizuno, S. Biogenic Peptides and Their Potential Use. Curr. Pharm. Des. 2003, 9, 1345–1355. [Google Scholar] [CrossRef]
- López Expósito, I.; Recio, I. Antibacterial activity of peptides and folding variants from milk proteins. Int. Dairy J. 2006, 16, 1294–1305. [Google Scholar] [CrossRef]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Lakshmaiah Narayana, J.; Chen, J.-Y. Antimicrobial peptides: Possible anti-infective agents. Peptides 2015, 72, 88–94. [Google Scholar] [CrossRef]
- Montone, C.M.; Capriotti, A.L.; Cerrato, A.; Antonelli, M.; La Barbera, G.; Piovesana, S.; Laganà, A.; Cavaliere, C. Identification of bioactive short peptides in cow milk by high-performance liquid chromatography on C18 and porous graphitic carbon coupled to high-resolution mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 3395–3404. [Google Scholar] [CrossRef]
- Piovesana, S.; Montone, C.M.; Cavaliere, C.; Crescenzi, C.; La Barbera, G.; Laganà, A.; Capriotti, A.L. Sensitive untargeted identification of short hydrophilic peptides by high performance liquid chromatography on porous graphitic carbon coupled to high resolution mass spectrometry. J. Chromatogr. A 2019, 1590, 73–79. [Google Scholar] [CrossRef]
- Piovesana, S.; Capriotti, A.L.; Cerrato, A.; Crescenzi, C.; La Barbera, G.; Laganà, A.; Montone, C.M.; Cavaliere, C. Graphitized Carbon Black Enrichment and UHPLC-MS/MS Allow to Meet the Challenge of Small Chain Peptidomics in Urine. Anal. Chem. 2019, 91, 11474–11481. [Google Scholar] [CrossRef] [PubMed]
- Cerrato, A.; Aita, S.E.; Capriotti, A.L.; Cavaliere, C.; Montone, C.M.; Laganà, A.; Piovesana, S. A new opening for the tricky untargeted investigation of natural and modified short peptides. Talanta 2020, 219, 121262. [Google Scholar] [CrossRef]
- Montone, C.M.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Zenezini Chiozzi, R.; Laganà, A. Characterization of antioxidant and angiotensin-converting enzyme inhibitory peptides derived from cauliflower by-products by multidimensional liquid chromatography and bioinformatics. J. Funct. Foods 2018, 44, 40–47. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, H.J.; Yoon, I.S.; Lee, G.-W.; Kim, J.-S.; Heu, M.S. Protein functionality of concentrates prepared from yellowfin tuna (Thunnus albacares) roe by cook-dried process. Food Sci. Biotechnol. 2016, 25, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Zenezini Chiozzi, R.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Laganà, A. Identification of three novel angiotensin-converting enzyme inhibitory peptides derived from cauliflower by-products by multidimensional liquid chromatography and bioinformatics. J. Funct. Foods 2016, 27, 262–273. [Google Scholar] [CrossRef]
- Montone, C.M.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Zenezini Chiozzi, R.; Laganà, A. Peptidomic strategy for purification and identification of potential ACE-inhibitory and antioxidant peptides in Tetradesmus obliquus microalgae. Anal. Bioanal. Chem. 2018, 410, 3573–3586. [Google Scholar] [CrossRef]
- Zenezini Chiozzi, R.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Samperi, R.; Laganà, A. Purification and identification of endogenous antioxidant and ACE-inhibitory peptides from donkey milk by multidimensional liquid chromatography and nanoHPLC-high resolution mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 5657–5666. [Google Scholar] [CrossRef]
- Esmaeilpour, M.; Ehsani, M.R.; Aminlari, M.; Shekarforoush, S.; Hoseini, E. Antimicrobial activity of peptides derived from enzymatic hydrolysis of goat milk caseins. Comp. Clin. Path. 2016, 25, 599–605. [Google Scholar] [CrossRef]
- Feßler, A.T.; Schug, A.R.; Geber, F.; Scholtzek, A.D.; Merle, R.; Brombach, J.; Hensel, V.; Meurer, M.; Michael, G.B.; Reinhardt, M.; et al. Development and evaluation of a broth macrodilution method to determine the biocide susceptibility of bacteria. Vet. Microbiol. 2018, 223, 59–64. [Google Scholar] [CrossRef]
- Proroga, Y.T.R.; Mancusi, A.; Peruzy, M.F.; Carullo, M.R.; Montone, A.M.I.; Fulgione, A.; Capuano, F. Characterization of Salmonella Typhimurium and its monophasic variant 1,4, [5],12:i:- isolated from different sources. Folia Microbiol. (Praha) 2019, 64, 711–718. [Google Scholar] [CrossRef]
- Niedermeyer, T.H.J.; Strohalm, M. mMass as a Software Tool for the Annotation of Cyclic Peptide Tandem Mass Spectra. PLoS ONE 2012, 7, e44913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel, F.B.; Cermeño, M.; Kleekayai, T.; Harnedy, P.A.; FitzGerald, R.J.; Alves, R.C.; Oliveira, M.B.P.P. Effect of in vitro simulated gastrointestinal digestion on the antioxidant activity of the red seaweed Porphyra dioica. Food Res. Int. 2020, 136, 109309. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive Peptides from Muscle Sources: Meat and Fish. Nutrients 2011, 3, 765–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezeshk, S.; Ojagh, S.M.; Rezaei, M.; Shabanpour, B. Fractionation of Protein Hydrolysates of Fish Waste Using Membrane Ultrafiltration: Investigation of Antibacterial and Antioxidant Activities. Probiotics Antimicrob. Proteins 2019, 11, 1015–1022. [Google Scholar] [CrossRef]
- Salampessy, J.; Phillips, M.; Seneweera, S.; Kailasapathy, K. Release of antimicrobial peptides through bromelain hydrolysis of leatherjacket (Meuchenia sp.) insoluble proteins. Food Chem. 2010, 120, 556–560. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Combination Effects of Antimicrobial Peptides. Antimicrob. Agents Chemother. 2016, 60, 1717–1724. [Google Scholar] [CrossRef] [Green Version]
- Kolker, E.; Higdon, R.; Haynes, W.; Welch, D.; Broomall, W.; Lancet, D.; Stanberry, L.; Kolker, N. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. 2012, 40, D1093–D1099. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, A.L.; Cavaliere, C.; Foglia, P.; Piovesana, S.; Samperi, R.; Stampachiacchiere, S.; Laganà, A. Proteomic platform for the identification of proteins in olive (Olea europaea) pulp. Anal. Chim. Acta 2013, 800, 36–42. [Google Scholar] [CrossRef]
- Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Foglia, P.; Piovesana, S.; Samperi, R.; Laganà, A. Proteome investigation of the non-model plant pomegranate (Punica granatum L.). Anal. Bioanal. Chem. 2013, 405, 9301–9309. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Harris, F.; Dennison, S.; Phoenix, D. Anionic Antimicrobial Peptides from Eukaryotic Organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef]
- Holton, T.A.; Pollastri, G.; Shields, D.C.; Mooney, C. CPPpred: Prediction of cell penetrating peptides. Bioinformatics 2013, 29, 3094–3096. [Google Scholar] [CrossRef]
- Ramesh, S.; Govender, T.; Kruger, H.G.; de la Torre, B.G.; Albericio, F. Short AntiMicrobial Peptides (SAMPs) as a class of extraordinary promising therapeutic agents. J. Pept. Sci. 2016, 22, 438–451. [Google Scholar] [CrossRef]
- Blondelle, S.E.; Houghten, R.A. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry 1992, 31, 12688–12694. [Google Scholar] [CrossRef]
- Deslouches, B.; Phadke, S.M.; Lazarevic, V.; Cascio, M.; Islam, K.; Montelaro, R.C.; Mietzner, T.A. De Novo Generation of Cationic Antimicrobial Peptides: Influence of Length and Tryptophan Substitution on Antimicrobial Activity. Antimicrob. Agents Chemother. 2005, 49, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Hans, D.; Young, P.; Fairlie, D. Current Status Of Short Synthetic Peptides As Vaccines. Med. Chem. (Los Angel.) 2006, 2, 627–646. [Google Scholar] [CrossRef]
- Olagunju, A.I.; Omoba, O.S.; Enujiugha, V.N.; Alashi, A.M.; Aluko, R.E. Antioxidant properties, ACE/renin inhibitory activities of pigeon pea hydrolysates and effects on systolic blood pressure of spontaneously hypertensive rats. Food Sci. Nutr. 2018, 6, 1879–1889. [Google Scholar] [CrossRef]
- Boachie, R.T.; Okoro, F.L.; Imai, K.; Sun, L.; Elom, S.O.; Nwankwo, J.O.; Ejike, C.E.C.C.; Udenigwe, C.C. Enzymatic release of dipeptidyl peptidase-4 inhibitors (gliptins) from pigeon pea (Cajanus cajan) nutrient reservoir proteins: In silico and in vitro assessments. J. Food Biochem. 2019, 43. [Google Scholar] [CrossRef] [PubMed]
- Goumon, Y.; Lugardon, K.; Kieffer, B.; Lefèvre, J.-F.; Van Dorsselaer, A.; Aunis, D.; Metz-Boutigue, M.-H. Characterization of Antibacterial COOH-terminal Proenkephalin-A-derived Peptides (PEAP) in Infectious Fluids. J. Biol. Chem. 1998, 273, 29847–29856. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| C18 Extract | GCB Extract | |||
|---|---|---|---|---|
| Concentration (mg mL−1) | MIC (cfu mL−1) | OD600 | MIC (cfu mL−1) | OD600 |
| 7.0 | 0 | 0.000 | 0 | 0.000 |
| 3.5 | 0 | 0.000 | 0 | 0.000 |
| 1.0 | 0 | 0.000 | 1 | 0.021 |
| 0.5 | 3 | 0.018 | 3 | 0.023 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerrato, A.; Capriotti, A.L.; Capuano, F.; Cavaliere, C.; Montone, A.M.I.; Montone, C.M.; Piovesana, S.; Zenezini Chiozzi, R.; Laganà, A. Identification and Antimicrobial Activity of Medium-Sized and Short Peptides from Yellowfin Tuna (Thunnus albacares) Simulated Gastrointestinal Digestion. Foods 2020, 9, 1185. https://doi.org/10.3390/foods9091185
Cerrato A, Capriotti AL, Capuano F, Cavaliere C, Montone AMI, Montone CM, Piovesana S, Zenezini Chiozzi R, Laganà A. Identification and Antimicrobial Activity of Medium-Sized and Short Peptides from Yellowfin Tuna (Thunnus albacares) Simulated Gastrointestinal Digestion. Foods. 2020; 9(9):1185. https://doi.org/10.3390/foods9091185
Chicago/Turabian StyleCerrato, Andrea, Anna Laura Capriotti, Federico Capuano, Chiara Cavaliere, Angela Michela Immacolata Montone, Carmela Maria Montone, Susy Piovesana, Riccardo Zenezini Chiozzi, and Aldo Laganà. 2020. "Identification and Antimicrobial Activity of Medium-Sized and Short Peptides from Yellowfin Tuna (Thunnus albacares) Simulated Gastrointestinal Digestion" Foods 9, no. 9: 1185. https://doi.org/10.3390/foods9091185