A Novel Fluorescence Aptasensor Based on Magnetic Beads/Gold Nanoparticles/DNA-Stabilized Silver Nanoclusters for Detection of Salmonella Typhimurium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation
2.3. Bacterial Strains and Cultivation Conditions
2.4. Preparation of SMBs-cDNA1 Complex
2.5. Production of the AuNPs and AuNPs-cDNA2 Probe
2.6. Synthesis of DNA-AgNCs
2.7. Fluorescence Aptasensor Assay of S. Typhimurium
2.8. Sensitivity Assessment in Milk Samples
3. Results and Discussion
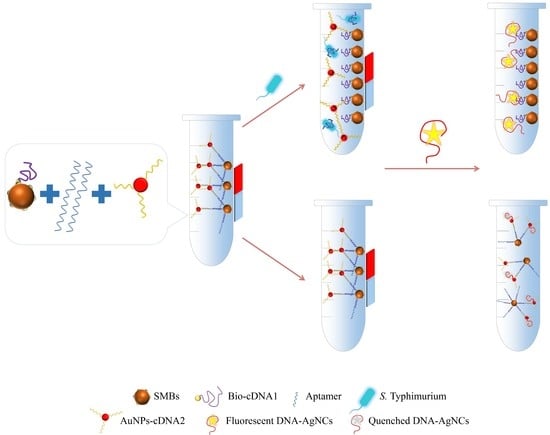
3.1. Principle of Detection Method
3.2. Characterization of AuNPs and DNA-AgNCs
3.3. Feasibility Testing for Proposed Strategy
3.4. Optimization of Trial Conditions
3.5. Sensitivity Detection for S. Typhimurium
3.6. Sensitivity Test in Artificially Contaminated Milk Samples
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Srisa-Art, M.; Boehle, K.E.; Geiss, B.J.; Henry, C.S. Highly Sensitive and Rapid Detection of Salmonella typhimurium Using a Colorimetric Paper-Based Analytical Device Coupled with Immunomagnetic Separation. Anal. Chem. 2018, 90, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; McWhorter, A.R.; Andrews, D.M.; Underwood, G.J.; Chousalkar, K.K. Challenges in Vaccinating Layer Hens against Salmonella Typhimurium. Vaccines 2020, 8, 696. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, L.; Cai, G.; Liu, N.; Liao, M.; Li, Y.; Zhang, X.; Lin, J. A microfluidic biosensor for online and sensitive detection of Salmonella typhimurium using fluorescence labeling and smartphone video processing. Biosens. Bioelectron. 2019, 140, 111333. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Hammerl, J.A.; Boone, I.; Jansen, W.; Fohler, S.; Klein, G.; Dieckmann, R.; Al Dahouk, S. Overview of validated alternative methods for the detection of foodborne bacterial pathogens. Trends Food Sci. Technol. 2017, 62, 113–118. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, R.; Sun, W.; Lv, L.; Guo, Z. Ochratoxin A detection platform based on signal amplification by Exonuclease III and fluorescence quenching by gold nanoparticles. Sens. Actuators B Chem. 2018, 255, 1640–1645. [Google Scholar] [CrossRef]
- Yu, X.; Chen, F.; Wang, R.; Li, Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. J. Biotechnol. 2018, 266, 39–49. [Google Scholar] [CrossRef]
- Ma, X.; Song, L.; Zhou, N.; Xia, Y.; Wang, Z. A novel aptasensor for the colorimetric detection of S. typhimurium based on gold nanoparticles. Int. J. Food Microbiol. 2017, 245, 1–5. [Google Scholar] [CrossRef]
- Srinivasan, S.; Ranganathan, V.; DeRosa, M.C.; Murari, B.M. Label-free aptasensors based on fluorescent screening assays for the detection of Salmonella typhimurium. Anal. Biochem. 2018, 559, 17–23. [Google Scholar] [CrossRef]
- Ranjbar, S.; Shahrokhian, S.; Nurmohammadi, F. Nanoporous gold as a suitable substrate for preparation of a new sensitive electrochemical aptasensor for detection of Salmonella typhimurium. Sens. Actuators B Chem. 2018, 255, 1536–1544. [Google Scholar] [CrossRef]
- Chen, Q.; Gao, R.; Jia, L. Enhancement of the peroxidase-like activity of aptamers modified gold nanoclusters by bacteria for colorimetric detection of Salmonella typhimurium. Talanta 2021, 221, 121476. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhu, X.; Zhou, X.; Khusbu, F.Y.; Ma, C. Recent advances in the bioanalytical and biomedical applications of DNA-templated silver nanoclusters. TrAC Trends Anal. Chem. 2020, 124, 115786. [Google Scholar] [CrossRef]
- Zou, R.; Zhang, F.; Chen, C.; Cai, C. DNA-programming multicolor silver nanoclusters for sensitively simultaneous detection of two HIV DNAs. Sens. Actuators B Chem. 2019, 296, 126608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gao, G.; Ding, Y.; Deng, C.; Xiang, J.; Wu, H. A fluorescent aptasensor for the femtomolar detection of epidermal growth factor receptor-2 based on the proximity of G-rich sequences to Ag nanoclusters. Talanta 2019, 199, 238–243. [Google Scholar] [CrossRef]
- Saraswathi, S.K.; Vittala, S.K.; Manayani, M.K.; Joseph, J. Sequence programmed DNA three-way junctions for templated assembly of fluorescent silver nanoclusters. J. Photochem. Photobiol. B Biol. 2020, 207, 111886. [Google Scholar] [CrossRef]
- Feng, X.; Han, T.; Xiong, Y.; Wang, S.; Dai, T.; Chen, J.; Zhang, X.; Wang, G. Plasmon-Enhanced Electrochemiluminescence of Silver Nanoclusters for microRNA Detection. ACS Sens. 2019, 4, 1633–1640. [Google Scholar] [CrossRef]
- Lee, C.Y.; Lin, S.W.; Wu, Y.H.; Hsieh, Y.Z. Combining DNA-stabilized silver nanocluster synthesis with exonuclease III amplification allows label-free detection of coralyne. Anal. Chim. Acta 2018, 1042, 86–92. [Google Scholar] [CrossRef]
- Lee, S.Y.; Fazlina, N.; Tye, G.J. DNA-templated silver nanocluster for live-intracellular FOXP3 detection. Anal. Biochem. 2019, 581, 113352. [Google Scholar] [CrossRef]
- Esmaelpourfarkhani, M.; Abnous, K.; Taghdisi, S.M.; Chamsaz, M. A novel turn-off fluorescent aptasensor for ampicillin detection based on perylenetetracarboxylic acid diimide and gold nanoparticles. Biosens. Bioelectron. 2020, 164, 112329. [Google Scholar] [CrossRef]
- Asnaashari, M.; Esmaeilzadeh Kenari, R.; Farahmandfar, R.; Taghdisi, S.M.; Abnous, K. Fluorescence quenching biosensor for acrylamide detection in food products based on double-stranded DNA and gold nanoparticles. Sens. Actuators B Chem. 2018, 265, 339–345. [Google Scholar] [CrossRef]
- Ma, J.L.; Yin, B.C.; Le, H.N.; Ye, B.C. Label-Free Detection of Sequence-Specific DNA Based on Fluorescent Silver Nanoclusters-Assisted Surface Plasmon-Enhanced Energy Transfer. ACS Appl. Mater. Interfaces 2015, 7, 12856–12863. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Cheng, Z.; Ma, H.; Li, Z.; Xue, N.; Wang, P. Label-Free Platform for MicroRNA Detection Based on the Fluorescence Quenching of Positively Charged Gold Nanoparticles to Silver Nanoclusters. Anal. Chem. 2018, 90, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, L.; Pang, L.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. A novel fluorescent platform of DNA-stabilized silver nanoclusters based on exonuclease III amplification-assisted detection of Salmonella Typhimurium. Anal. Chim. Acta 2021, 1181, 338903. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Ma, Y.; Zhang, A.; Zhang, L.; Zeng, L.; Liu, G. Disposable Nucleic Acid Biosensors Based on Gold Nanoparticle Probes and Lateral Flow Strip. Anal. Chem. 2009, 81, 1660–1668. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, X.; Fu, S.; Qin, X.; Yang, T.; Man, C.; Jiang, Y. A novel AuNPs colorimetric sensor for sensitively detecting viable Salmonella typhimurium based on dual aptamers. Food Control 2020, 115, 107281. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, H.; Li, W.; Dong, Y.; Chi, Y. A novel hybrid platform of g-C3N4 nanosheets /nucleic-acid-stabilized silver nanoclusters for sensing protein. Anal. Chim. Acta 2019, 1091, 112–118. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2017.
- Li, S.; Fu, Y.; Ma, X.; Zhang, Y. Label-free fluorometric detection of chymotrypsin activity using graphene oxide/nucleic-acid-stabilized silver nanoclusters hybrid materials. Biosens. Bioelectron. 2017, 88, 210–216. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, K.; Li, K.-B.; Shi, W.; Jia, W.-P.; Chen, X.; Sun, T.; Han, D.-M. A DNA-stabilized silver nanoclusters/graphene oxide-based platform for the sensitive detection of DNA through hybridization chain reaction. Biosens. Bioelectron. 2017, 91, 374–379. [Google Scholar] [CrossRef]
- Xue, N.; Wu, S.; Li, Z.; Miao, X. Ultrasensitive and label-free detection of ATP by using gold nanorods coupled with enzyme assisted target recycling amplification. Anal. Chim. Acta 2020, 1104, 117–124. [Google Scholar] [CrossRef]
- Lerga, T.M.; Skouridou, V.; Bermudo, M.C.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. Gold nanoparticle aptamer assay for the determination of histamine in foodstuffs. Microchim. Acta 2020, 187, 452. [Google Scholar] [CrossRef]
- Kimura-Suda, H.; Petrovykh, D.Y.; Tarlov, M.J.; Whitman, L.J. Base-Dependent Competitive Adsorption of Single-Stranded DNA on Gold. J. Am. Chem. Soc. 2003, 125, 9014–9015. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, R.; Chen, F.; Jiang, T.; Wang, H.; Slavik, M.; Wei, H.; Li, Y. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017, 221, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Q.; Han, L.; Du, S.; Yu, H.; Zhang, H. Rapid and sensitive detection of Salmonella typhimurium based on the photothermal effect of magnetic nanomaterials. Sens. Actuators B Chem. 2018, 268, 188–194. [Google Scholar] [CrossRef]
- Ma, X.; Xu, X.; Xia, Y.; Wang, Z. SERS aptasensor for Salmonella typhimurium detection based on spiny gold nanoparticles. Food Control 2018, 84, 232–237. [Google Scholar] [CrossRef]
- Hu, J.; Jiang, Y.Z.; Tang, M.; Wu, L.L.; Xie, H.Y.; Zhang, Z.L.; Pang, D.W. Colorimetric-Fluorescent-Magnetic Nanosphere-Based Multimodal Assay Platform for Salmonella Detection. Anal. Chem. 2019, 91, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, L.; Song, S.; Tang, L.; Kuang, H.; Xu, C. A highly sensitive ELISA and immunochromatographic strip for the detection of Salmonella typhimurium in milk samples. Sensors 2015, 15, 5281–5292. [Google Scholar] [CrossRef]








| Primer | Sequence (5′-3′) |
|---|---|
| Aptamer | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG |
| cDNA1 | biotin-CTGTCATAATGTCAAGTCC |
| cDNA2 | GTCGGGTGACGCCGCCATA-SH |
| DNA | AGTGGAAAAACCCCCCCCCCCC |
| Type of Sensor | Detection Limit (cfu/mL) | Linear Range (cfu/mL) | Detection Time | References |
|---|---|---|---|---|
| Colorimetric aptasensor | 56 | 102 to107 | 31 h 15 min | [8] |
| Fluorescent aptasensor | 733 and 464 | 1530 to 96,938 | 30 min | [9] |
| Colorimetric aptasensor | 1 | 101 to 106 | 8 h | [11] |
| Gold nanoparticles (AuNPs) colorimetric sensor based on PCR assay | 33 | 3.3 × 101 to 3.3 × 106 | 3 h | [25] |
| Quartz crystal microbalance (QCM) biosensor | 7.9 × 103 | 7.9 × 102 to 7.9 × 106 | 1 h | [33] |
| Magnetic nanomaterial biosensor | 300 | 300 to 1000 | 1.5 h | [34] |
| Surface-enhanced Raman scattering (SERS) aptasensor | 4 | 101 to 105 | 14 h | [35] |
| Lateral flow immunoassay | 3.75 × 103 | 1.88 × 104 to 1.88 × 107 | 35 min | [36] |
| This method | 98 | 3.7 × 102 to 3.7 × 105 | 2 h 10 min |
| No. | Spiked (cfu/mL) | Found (cfu/mL) a | Recovery (%) | RSD (%, n = 3) |
|---|---|---|---|---|
| 1 | 5.2 × 104 | 5.65 × 104 | 108.7 | 4.91 |
| 2 | 5.2 × 105 | 5.08 × 105 | 97.7 | 1.88 |
| 3 | 5.2 × 106 | 4.83 × 106 | 92.9 | 2.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, S.; Yang, X.; Pang, L.; Cheng, S.; Song, D.; Qin, X.; Man, C.; Jiang, Y. A Novel Fluorescence Aptasensor Based on Magnetic Beads/Gold Nanoparticles/DNA-Stabilized Silver Nanoclusters for Detection of Salmonella Typhimurium. Foods 2022, 11, 595. https://doi.org/10.3390/foods11040595
Fu S, Yang X, Pang L, Cheng S, Song D, Qin X, Man C, Jiang Y. A Novel Fluorescence Aptasensor Based on Magnetic Beads/Gold Nanoparticles/DNA-Stabilized Silver Nanoclusters for Detection of Salmonella Typhimurium. Foods. 2022; 11(4):595. https://doi.org/10.3390/foods11040595
Chicago/Turabian StyleFu, Shiqian, Xinyan Yang, Lidong Pang, Shasha Cheng, Danliangmin Song, Xue Qin, Chaoxin Man, and Yujun Jiang. 2022. "A Novel Fluorescence Aptasensor Based on Magnetic Beads/Gold Nanoparticles/DNA-Stabilized Silver Nanoclusters for Detection of Salmonella Typhimurium" Foods 11, no. 4: 595. https://doi.org/10.3390/foods11040595