The Occurrence and Genomic Characteristics of mcr-1-Harboring Salmonella from Retail Meats and Eggs in Qingdao, China
Abstract
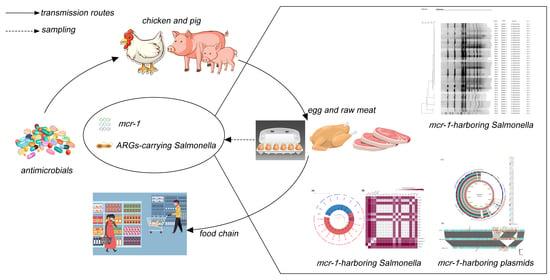
:1. Introduction
2. Materials and Methods
2.1. Isolation and Serotyping of Salmonella
2.2. Antimicrobial Susceptibility Testing and Identification of Resistance Genes
2.3. Pulsed-Field Gel Electrophoresis and Conjugation Experiments
2.4. Whole Genome Sequencing and Phylogenetic Analysis
2.5. Characterization of mcr-1-Bearing Plasmids
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Detection Rates and Serotypes of Salmonella in Raw Meats and Eggs
3.2. Antimicrobial Resistance Patterns and Genotypes
3.3. Conjugation and PFGE Analysis
3.4. Genomic Characteristics of mcr-1-Positive S. Typhimurium Isolates
3.5. Complete Sequence Analysis of mcr-1-Carrying Plasmids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of Novel Mobile Colistin Resistance Gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a Novel mcr-6 to mcr-9 Multiplex PCR and Assessment of mcr-1 to mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates From Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Wu, Z.; Wang, Y.; Zhang, R.; Zhou, H.W.; Wang, S.; Lei, L.; Li, M.; Cai, J.; Tyrrell, J.; et al. Heterogeneous and Flexible Transmission of mcr-1 in Hospital-Associated Escherichia coli. MBio 2018, 9, e00943-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, N.L.; Pimenta, R.L.; de Melo, D.A.; Nolan, L.K.; de Souza, M.M.S.; Logue, C.M. mcr-1 Identified in Fecal Escherichia coli and Avian Pathogenic E. coli (APEC) From Brazil. Front. Microbiol. 2021, 12, 659613. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated Aquaculture Contributes to the Transfer of mcr-1 between Animals and Humans via the Aquaculture Supply Chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kuang, X.; Liu, J.; Sun, R.Y.; Li, X.P.; Sun, J.; Liao, X.P.; Liu, Y.H.; Yu, Y. Identification of the Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli Isolates From Migratory Birds in Guangdong, China. Front. Microbiol. 2021, 12, 755233. [Google Scholar] [CrossRef]
- Torres, R.T.; Cunha, M.V.; Araujo, D.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. Emergence of Colistin Resistance Genes (mcr-1) in Escherichia coli among Widely Distributed Wild Ungulates. Environ. Pollut. 2021, 291, 118136. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.H.; Wu, P.C.; Tang, S.L.; Chen, Y.C.; Cheng, M.F.; Huang, P.C.; Ko, W.C.; Wang, J.L. A Large Spatial Survey of Colistin-Resistant Gene mcr-1-Carrying E. coli in Rivers across Taiwan. Microorganisms 2021, 9, 722. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.E.; Tansawai, U.; Andrey, D.O.; Wang, S.; Wang, Y.; Sands, K.; Kiddee, A.; Assawatheptawee, K.; Bunchu, N.; Hassan, B.; et al. Environmental Dissemination of mcr-1 Positive Enterobacteriaceae by Chrysomya spp. (Common Blowfly): An Increasing Public Health Risk. Environ. Int. 2019, 122, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.T.; Li, X.; Zhang, Q.; Shan, H.; Zou, M.; Song, F.J. Colistin-Resistant mcr-Positive Enterobacteriaceae in Fresh Vegetables, an Increasing Infectious Threat in China. Int. J. Antimicrob. Agents 2019, 54, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, H.; Kang, H.S.; Kim, Y.; Kim, M.; Kwak, H.; Ryu, S. Prevalence and Genetic Characterization of mcr-1-Positive Escherichia coli Isolated from Retail Meats in South Korea. J. Microbiol. Biotechnol. 2020, 30, 1862–1869. [Google Scholar] [CrossRef] [PubMed]
- Besser, J.M. Salmonella Epidemiology: A Whirlwind of Change. Food Microbiol. 2018, 71, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S. Salmonella in the Pork Production Chain and Its Impact on Human Health in the European Union. Epidemiol. Infect. 2017, 145, 1513–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Ed-Dra, A.; Pan, H.; Dong, C.; Jia, C.; Yue, M. Genomic Investigation of Salmonella Isolates Recovered From a Pig Slaughtering Process in Hangzhou, China. Front. Microbiol. 2021, 12, 704636. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zeng, M.; Xu, J.; Zhou, H.; Gu, B.; Li, Z.; Jin, H.; Wang, X.; Zhang, W.; Hu, Y.; et al. Epidemiologic and Genomic Insights on mcr-1-Harbouring Salmonella from Diarrhoeal Outpatients in Shanghai, China, 2006-2016. EBioMedicine 2019, 42, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Zhang, J.; Gu, Z.; Li, R.; Chan, E.W.; Yan, M.; Wu, C.; Xu, X.; Chen, S. Prevalence and Molecular Characterization of mcr-1-Positive Salmonella Strains Recovered from Clinical Specimens in China. Antimicrob. Agents Chemother. 2017, 61, e02471-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortini, D.; Owczarek, S.; Dionisi, A.M.; Lucarelli, C.; Arena, S.; Carattoli, A.; Enter-Net Italia Colistin Resistance Study Group; Villa, L.; Garcia-Fernandez, A. Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics 2022, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fanning, S.; Gan, X.; Liu, C.; Nguyen, S.; Wang, M.; Wang, W.; Jiang, T.; Xu, J.; Li, F. Salmonella Harbouring the mcr-1 Gene Isolated from Food in China between 2012 and 2016. J. Antimicrob. Chemother. 2019, 74, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Rau, R.B.; de Lima-Morales, D.; Wink, P.L.; Ribeiro, A.R.; Martins, A.F.; Barth, A.L. Emergence of mcr-1 Producing Salmonella enterica Serovar Typhimurium from Retail Meat: First Detection in Brazil. Foodborne Pathog. Dis. 2018, 15, 58–59. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Wan, F.; Yu, X.; Zheng, B.; Chen, Y.; Gong, C.; Fu, H.; Xiao, Y.; Li, L. MDR Salmonella enterica Serovar Typhimurium ST34 Carrying mcr-1 Isolated from Cases of Bloodstream and Intestinal Infection in Children in China. J. Antimicrob. Chemother. 2020, 75, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fanning, S.; Nguyen, S.V.; Wang, W.; Liu, C.; Cui, X.; Dong, Y.; Gan, X.; Xu, J.; Li, F. Emergence of a Salmonella enterica Serovar Typhimurium ST34 isolate, CFSA629, Carrying a Novel mcr-1.19 Variant Cultured from Egg in China. J. Antimicrob. Chemother. 2021, 76, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-N.; Chen, J.-S. Food Safety Monitoring and Surveillance in China: Past, Present and Future. Food Control 2018, 90, 429–439. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Zheng, Z.; Chen, K.; Xie, M.; Chan, E.W.; Geng, S.; Chen, S. Molecular Characterization of Escherichia coli Isolates Carrying mcr-1, fosA3, and Extended-Spectrum-beta-Lactamase Genes from Food Samples in China. Antimicrob. Agents Chemother. 2017, 61, e00064-17. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Change, J.; Luo, Y.; Jiang, H.; Liu, C.; Xiao, X.; Ji, X.; Yang, H. Prevalence and Characteristics of the mcr-1 Gene in Retail Meat Samples in Zhejiang Province, China. J. Microbiol. 2022, 60, 610–619. [Google Scholar] [CrossRef]
- Liu, X.; Geng, S.; Chan, E.W.; Chen, S. Increased Prevalence of Escherichia coli Strains from Food Carrying blaNDM and mcr-1-Bearing Plasmids that Structurally Resemble Those of Clinical Strains, China, 2015 to 2017. Eurosurveillance 2019, 24, 1800113. [Google Scholar] [CrossRef] [Green Version]
- Lyu, N.; Feng, Y.; Pan, Y.; Huang, H.; Liu, Y.; Xue, C.; Zhu, B.; Hu, Y. Genomic Characterization of Salmonella enterica Isolates From Retail Meat in Beijing, China. Front. Microbiol. 2021, 12, 636332. [Google Scholar] [CrossRef]
- Hu, Y.; Nguyen, S.V.; Wang, W.; Gan, X.; Dong, Y.; Liu, C.; Cui, X.; Xu, J.; Li, F.; Fanning, S. Antimicrobial Resistance and Genomic Characterization of Two mcr-1-Harboring Foodborne Salmonella Isolates Recovered in China, 2016. Front. Microbiol. 2021, 12, 636284. [Google Scholar] [CrossRef]
- Agbaje, M.; Ayo-Ajayi, P.; Kehinde, O.; Omoshaba, E.; Dipeolu, M.; Fasina, F.O. Salmonella Characterization in Poultry Eggs Sold in Farms and Markets in Relation to Handling and Biosecurity Practices in Ogun State, Nigeria. Antibiotics 2021, 10, 773. [Google Scholar] [CrossRef]
- GB 4789.4-2016; National Food Safety Standard Food Microbiological Examination: Salmonella. NHFPC: Beijing, China, 2016.
- Ferretti, R.; Mannazzu, I.; Cocolin, L.; Comi, G.; Clementi, F. Twelve-Hour PCR-Based Method for Detection of Salmonella spp. in Food. Appl. Environ. Microbiol. 2001, 67, 977–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Xu, X.; Li, K.; Liu, B.; Yue, T. Identification of Salmonella enterica Typhimurium and Variants Using a Novel Multiplex PCR Assay. Food Control 2016, 65, 152–159. [Google Scholar] [CrossRef]
- Xiong, D.; Song, L.; Tao, J.; Zheng, H.; Zhou, Z.; Geng, S.; Pan, Z.; Jiao, X. An Efficient Multiplex PCR-Based Assay as a Novel Tool for Accurate Inter-Serovar Discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin. Front. Microbiol. 2017, 8, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Frye, J.G.; Hu, J.; Fedorka-Cray, P.J.; Gautom, R.; Boyle, D.S. Multiplex PCR-Based Method for Identification of Common Clinical Serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 2006, 44, 3608–3615. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. In CLSI Document M100-S25; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0. Växjö, Sweden: European Committee on Antimicrobial Susceptibility Testing. 2019. Available online: http://www.eucast.org/clinical_breakpoints (accessed on 1 January 2019).
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.T.; Song, F.J.; Zou, M. Characterization of Highly Prevalent Plasmids Coharboring mcr-1, oqxAB, and blaCTX-M and Plasmids Harboring oqxAB and blaCTX-M in Escherichia coli Isolates from Food-Producing Animals in China. Microb. Drug Resist. 2019, 25, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Sriskandan, S.; Woodford, N.; Hopkins, K.L. High Prevalence of 16S rRNA Methyltransferases among Carbapenemase-Producing Enterobacteriaceae in the UK and Ireland. Int. J. Antimicrob. Agents 2018, 52, 278–282. [Google Scholar] [CrossRef]
- Gautom, R.K. Rapid Pulsed-Field Gel Electrophoresis Protocol for Typing of Escherichia coli O157:H7 and other Gram-Negative Organisms in 1 Day. J. Clin. Microbiol. 1997, 35, 2977–2980. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisi, A.; Crump, J.A.; Glass, K.; Howden, B.P.; Furuya-Kanamori, L.; Vilkins, S.; Gray, D.J.; Kirk, M.D. Health Outcomes from Multidrug-Resistant Salmonella Infections in High-Income Countries: A Systematic Review and Meta-Analysis. Foodborne Pathog. Dis. 2018, 15, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, X.; Hou, W.; Wang, H.; Paoli, G.C.; Shi, X. Incidence and Characterization of Salmonella Isolates From Raw Meat Products Sold at Small Markets in Hubei Province, China. Front. Microbiol. 2019, 10, 2265. [Google Scholar] [CrossRef]
- Yin, X.; M’Ikanatha, N.M.; Nyirabahizi, E.; McDermott, P.F.; Tate, H. Antimicrobial Resistance in Non-Typhoidal Salmonella from Retail Poultry Meat by Antibiotic Usage-Related Production Claims—United States, 2008–2017. Int. J. Food Microbiol. 2021, 342, 109044. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Liu, S.; Zhang, Y.; Chen, C.; Xu, M.; Zhu, Y.; Chen, B.; Zhou, W.; Cui, S.; et al. Prevalence, Antimicrobial Resistance, and Genotype Diversity of Salmonella Isolates Recovered from Retail Meat in Hebei Province, China. Int. J. Food Microbiol. 2022, 364, 109515. [Google Scholar] [CrossRef]
- Haubert, L.; Maia, D.S.V.; Rauber Wurfel, S.F.; Vaniel, C.; da Silva, W.P. Virulence Genes and Sanitizers Resistance in Salmonella Isolates from Eggs in Southern Brazil. J. Food Sci. Technol. 2022, 59, 1097–1103. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Mashak, Z.; Ghadimianazar, A. Prevalence and Antimicrobial Resistance of Salmonella Serotypes Isolated from Retail Chicken Meat and Giblets in Iran. J. Infect. Dev. Ctries. 2015, 9, 463–469. [Google Scholar] [CrossRef] [Green Version]
- Krishnasamy, V.; Otte, J.; Silbergeld, E. Antimicrobial Use in Chinese Swine and Broiler Poultry Production. Antimicrob. Resist. Infect. Control 2015, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Elbediwi, M.; Beibei, W.; Pan, H.; Jiang, Z.; Biswas, S.; Li, Y.; Yue, M. Genomic Characterization of mcr-1-Carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China. Front. Bioeng. Biotechnol. 2020, 8, 663. [Google Scholar] [CrossRef]
- Zhang, H.; Xiang, Y.; Huang, Y.; Liang, B.; Xu, X.; Xie, J.; Du, X.; Yang, C.; Liu, H.; Liu, H.; et al. Genetic Characterization of mcr-1-Positive Multidrug-Resistant Salmonella enterica Serotype Typhimurium Isolated From Intestinal Infection in Children and Pork Offal in China. Front. Microbiol. 2021, 12, 774797. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Fang, L.X.; Song, J.Q.; Xia, J.; Huo, W.; Fang, J.T.; Liao, X.P.; Liu, Y.H.; Feng, Y.; Sun, J. Clonal Spread of mcr-1 in PMQR-Carrying ST34 Salmonella Isolates from Animals in China. Sci. Rep. 2016, 6, 38511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, N.; Sekizuka, T.; Tamamura, Y.; Tanaka, K.; Barco, L.; Izumiya, H.; Kusumoto, M.; Hinenoya, A.; Yamasaki, S.; Iwata, T.; et al. Phylogenetic Characterization of Salmonella enterica Serovar Typhimurium and Its Monophasic Variant Isolated from Food Animals in Japan Revealed Replacement of Major Epidemic Clones in the Last 4 Decades. J. Clin. Microbiol. 2018, 56, e01758-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, L.; Wang, J.; Gao, Y.; Liu, Y.; Doi, Y.; Wu, R.; Zeng, Z.; Liang, Z.; Liu, J.H. mcr-1-Harboring Salmonella enterica Serovar Typhimurium Sequence Type 34 in Pigs, China. Emerg. Infect. Dis. 2017, 23, 291–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, R.; Card, R.M.; Nunez-Garcia, J.; Mendonca, N.; da Silva, G.J.; Anjum, M.F. Multidrug-Resistant Salmonella enterica Isolated from Food Animal and Foodstuff May Also Be Less Susceptible to Heavy Metals. Foodborne Pathog. Dis. 2019, 16, 166–172. [Google Scholar] [CrossRef]




| Sample Types | Isolation Rates of Salmonella (%) | |
|---|---|---|
| Small Markets | Large Supermarkets | |
| Raw meat samples (n = 177) | - | 17.5 (31/177) |
| Chicken meat (n = 146) | - | 17.8 (26/146) |
| Pork (n = 31) | - | 16.1 (5/31) |
| Fresh eggs (n = 130) | 24.2 (16/66) | 0.0 (0/64) |
| Origins | Ratio of Different Serotypes (%) | ||||
|---|---|---|---|---|---|
| Enteritidis | Typhimurium | Braenderup | Newport | Not Identified | |
| Meat (n = 36) | 36.1 (13/36) | 8.3 (3/36) | 33.3 (12/36) | 5.6 (2/36) | 16.7 (6/36) |
| Chicken meat | 43.3 (13/30) | 3.3 (1/30) | 30.0 (9/30) | 6.7 (2/30) | 16.7 (5/30) |
| Pork | 0.0 (0/6) | 33.3 (2/6) | 50.0 (3/6) | 0.0 (0/6) | 16.7 (1/6) |
| Fresh eggs (n = 40) | 0.0 (0/40) | 100.0 (40/40) | 0.0 (0/40) | 0.0 (0/40) | 0.0 (0/40) |
| Total (n = 76) | 17.1 (13/76) | 56.6 (43/76) | 15.8 (12/76) | 2.6 (2/76) | 7.9 (6/76) |
| Antimicrobial Agents | Resistance Rates (%) | ||
|---|---|---|---|
| Retail Meats (n = 36) | Fresh Eggs (n = 40) | Total (n = 76) | |
| β-lactams | |||
| ampicillin | 55.6 (20/36) a | 100.0 (40/40) b | 78.9 (60/76) |
| ceftiofur | 8.3 (3/36) a | 2.5 (1/40) a | 5.3 (4/76) |
| cefotaxime | 8.3 (3/36) a | 0.0 (0/40) a | 3.9 (3/76) |
| meropenem | 0.0 (0/36) | 0.0 (0/40) | 0.0 (0/76) |
| Quinolones | |||
| nalidixic acid | 55.6 (20/36) a | 100.0 (40/40) b | 78.9 (60/76) |
| enrofloxacin | 8.3 (3/36) a | 0.0 (0/40) a | 3.9 (3/76) |
| ciprofloxacin | 8.3 (3/36) a | 5.0 (2/40) a | 6.6 (5/76) |
| levofloxacin | 5.6 (2/36) a | 0.0 (0/40) a | 2.6 (2/76) |
| Tetracyclines | |||
| tetracycline | 36.1 (13/36) a | 0.0 (0/40) b | 17.1 (13/76) |
| doxycycline | 36.1 (13/36) a | 0.0 (0/40) b | 17.1 (13/76) |
| tigecycline | 0.0 (0/36) | 0.0 (0/40) | 0.0 (0/76) |
| Aminoglycosides | |||
| streptomycin | 44.4 (16/36) a | 100.0 (40/40) b | 73.7 (56/76) |
| kanamycin | 25.0 (9/36) a | 92.5 (37/40) b | 60.5 (46/76) |
| gentamicin | 33.3 (12/36) a | 97.5 (39/40) b | 67.1 (51/76) |
| amikacin | 2.8 (1/36) a | 0.0 (0/40) a | 1.3 (1/76) |
| Lipopeptides | |||
| colistin | 0.0 (0/36) a | 100.0 (40/40) b | 52.6 (40/76) |
| Others | |||
| fosfomycin | 8.3 (3/36) a | 2.5 (1/40) a | 5.3 (4/76) |
| ≥3 (MDR) | 55.6 (20/36) a | 97.5 (39/40) b | 77.6 (59/76) |
| ≥4 | 27.8 (10/36) a | 97.5 (39/40) b | 64.5 (49/76) |
| ≥5 | 5.6 (2/36) a | 2.5 (1/40) a | 3.9 (3/76) |
| Isolates | Egg Brands | Markets | Resistance Genes | Replicon Type |
|---|---|---|---|---|
| S46L1 | Brand 5 | Market 3 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S58L2 | Brand 7 | Market 3 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, aph(3′)-Ia, aadA1, aadA2b, sul1, sul2, sul3, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y), oqxAB, floR, dfrA12, cmlA1 | IncHI2/HI2A |
| S34L1 | Brand 2 | Market 1 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S34L2 | Brand 2 | Market 1 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S34L3 | Brand 2 | Market 1 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S37L1 | Brand 3 | Market 2 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S37L3 | Brand 3 | Market 2 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S47L1 | Brand 5 | Market 3 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S54L1 | Brand 6 | Market 3 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
| S58L1 | Brand 7 | Market 3 | mcr-1, aph(4)-Ia, aac(6′)-Ib-cr, aac(3)-IV, aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, sul1, sul2, arr-3, blaOXA-1, blaTEM-1B, catB3, gyrA (D87Y) | IncHI2/HI2A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Gu, X.; Zhang, L.; Liu, Y.; Li, Y.; Zou, M.; Liu, B. The Occurrence and Genomic Characteristics of mcr-1-Harboring Salmonella from Retail Meats and Eggs in Qingdao, China. Foods 2022, 11, 3854. https://doi.org/10.3390/foods11233854
Li C, Gu X, Zhang L, Liu Y, Li Y, Zou M, Liu B. The Occurrence and Genomic Characteristics of mcr-1-Harboring Salmonella from Retail Meats and Eggs in Qingdao, China. Foods. 2022; 11(23):3854. https://doi.org/10.3390/foods11233854
Chicago/Turabian StyleLi, Changan, Xiulei Gu, Liping Zhang, Yuqing Liu, Yan Li, Ming Zou, and Baotao Liu. 2022. "The Occurrence and Genomic Characteristics of mcr-1-Harboring Salmonella from Retail Meats and Eggs in Qingdao, China" Foods 11, no. 23: 3854. https://doi.org/10.3390/foods11233854