Comparative Evaluation of Quality and Metabolite Profiles in Meju Using Starter Cultures of Bacillus velezensis and Aspergillus oryzae
Abstract
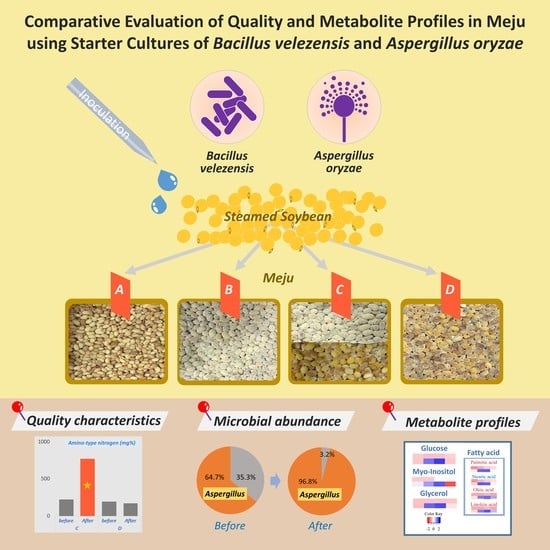
:1. Introduction
2. Materials and Methods
2.1. Manufacturing of the Modified Meju
2.2. Physicochemical Properties Analysis
2.3. Total Viable Counts and MiSeq Metagenomic Sequencing
2.4. Microbial Community Metagenome Using PICRUSt Based on 16S rRNA Sequencing Data
2.5. Volatile Analysis by GC-MS
2.6. Metabolite Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Physicochemical Properties of Modified Meju
3.2. Effect of Starter Inoculation Method on Changes in Bacterial and Fungal Communities
3.3. Volatile Components
3.4. Comparison of Metabolite Profiles
3.5. Heatmaps of Identified Metabolites
3.6. Predictive Functional Genes in Fungal and Bacterial Inoculants of Meju
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gil, N.Y.; Song, J.; Eom, J.S.; Park, S.Y.; Choi, H.S. Changes of physicochemical properties of Cheonggukjang prepared with various soybean cultivars and Bacillus subtilis HJ18-9. Korean J. Food Preserv. 2016, 23, 811–818. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.M.; Lim, H.J.; Kim, M.-S.; Kim, D.S.; Hwang, C.E.; Nam, S.H.; Joo, O.S.; Lee, B.W.; Kim, J.K.; Shin, E.C. Time course effects of fermentation on fatty acid and volatile compound profiles of Cheonggukjang using new soybean cultivars. J. Food Drug Anal. 2017, 25, 637–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omoni, A.O.; Aluko, R.E. Soybean foods and their benefits: Potential mechanisms of action. Nutr. Rev. 2005, 63, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.Y.; Choi, B.Y.; Park, S.Y.; Cho, Y.S.; Kim, S.Y. Physicochemical properties of Doenjang using grain type Meju fermented by Aspergillus oryzae and protease. Korean J. Food Preserv. 2017, 24, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.S.; Park, K.H.; Choi, K.J.; Won, S. Identification and isolation of zygomycetous fungi found on Maeju, a raw material of Korean traditional soysources. Kor. J. Mycol. 1993, 21, 172–187. (In Korean) [Google Scholar]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Microbial community dynamics during fermentation of doenjang-meju, traditional Korean fermented soybean. Int. J. Food Microbiol. 2014, 185, 112–120. [Google Scholar] [CrossRef]
- Jo, H.D.; Lee, H.A.; Jeong, S.J.; Kim, J.H. Purification and characterization of a major fibrinolytic enzyme from Bacillus amyloliquefaciens MJ5-41 isolated from Meju. J. Microbiol. Biotechnol. 2011, 21, 1166–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; Jeong, J.O.; Cho, S.H.; Jeong, D.Y.; Uhm, T.B. Antimicrobial and biogenic amine-degrading activity of Bacillus licheniformis SCK B11 isolated from traditionally fermented red pepper paste. Korean J. Microbiol. 2012, 48, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Ministry of Food and Drug Safety (MFD). Specifications and Standards for Foods; No. 2019-81; MFDS: Osong, Korea, 2019.
- Borriss, R.; Chen, X.H.; Rückert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7T and FZB42T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar]
- Kim, E.Y.; Kim, D.G.; Kim, Y.R.; Choi, S.Y.; Kong, I.S. Isolation and identification of halotolerant Bacillus sp. SJ-10 and characterization of its extracellular protease. Misainmurhag Hoiji. 2009, 45, 193–199. (In Korean) [Google Scholar]
- Chang, M.; Moon, S.H.; Chang, H.C. Isolation of Bacillus velezensis SSH100-10 with antifungal activity from Korean traditional soysauce and characterization of its antifungal compounds. Korean J. Food Preserv. 2012, 19, 757–766. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.H.; Park, Y.J.; Kim, I.C.; Chang, H.H. Isolation and characterization of Bacillus velezensis SS360-1 from seed soy sauce. Korean J. Community Living Sci. 2018, 29, 49–58. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, S.Y.; Cho, Y.S.; Gil, N.Y.; Choi, B.Y. Bacillus amyloliquefaciens NY12-2 Strain Having Antibiotic Activity against Bacillus cereus and Inhibitory Activity on Biogenic Amine and Compositions Thereof. Korean Patent No. 10-2000475, 10 July 2019. [Google Scholar]
- Association of Official Analytical Chemists (A.O.A.C.). Official Methods of Analysis, 15th ed.; A.O.A.C.: Washington, DC, USA, 1990. [Google Scholar]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 9, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, M.A. Isolation four species of Bacillus and tested activity against some filamentous fungi. Eur. J. Exp. Biol. 2013, 3, 495–498. [Google Scholar]
- Choi, J.; Kim, M.H.; Shon, M.Y.; Park, S.K.; Choi, S.-D.; Hong, U. Production and quality properties of capsule type Meju prepared with Rhizopus oligosporus. Korean J Food. Preserv. 2002, 9, 315–320. (In Korean) [Google Scholar]
- Liu, Y.; Teng, K.; Wang, T.; Dong, E.; Zhang, M.; Tao, Y.; Zhong, J. Antimicrobial Bacillus velezensis HC6: Production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 2020, 128, 242–254. [Google Scholar] [CrossRef] [Green Version]
- Thu, N.K.; Tanabe, Y.; Yoshida, M.; Matsuura, H.; Watanabe, M.M. Aerosakkonema funiforme gen. et sp. nov. (Oscillatoriales), a new gas-vacuolated oscillatorioid cyanobacterium isolated from a mesotrophic reservoir. Phycologia 2012, 51, 672–683. [Google Scholar] [CrossRef]
- Lee, L.; Heo, S.; Jeong, D.W. Fungal microbial community profiles of meju, solar salt, and doenjang using pyrosequencing. Microbiol. Biotechnol. Lett. 2019, 47, 354–358. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, M.Y.; Kim, K.S.; Lee, T.S. Volatile flavor components of soybean paste (doenjang) prepared from different types of strains. Korean J. Food Sci. Technol. 1994, 26, 255–260. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2019, 49, 1388–1395. [Google Scholar] [CrossRef]
- Seo, H.S.; Lee, S.; Singh, D.; Shin, H.W.; Cho, S.A.; Lee, C.H. Untargeted metabolite profiling for koji-fermentative bioprocess unravels the effects of varying substrate types and microbial inocula. Food Chem. 2018, 266, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H. Physicochemical and Functional Characteristics of Traditional Meju and Doenjang with Soybean in Different Seeding Times. Ph.D. Thesis, Seoul National University, Seoul, Korea, 2012. [Google Scholar]
- Lee, S.; Lee, S.; Singh, D.; Oh, J.Y.; Jeon, E.J.; Ryu, H.S.; Lee, D.W.; Kim, B.S.; Lee, C.H. Comparative evaluation of microbial diversity and metabolite profiles in doenjang, a fermented soybean paste, during the two different industrial manufacturing processes. Food Chem. 2017, 221, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Namgung, H.J.; Park, H.J.; Cho, I.H.; Choi, H.K.; Kwon, D.Y.; Shim, S.-M.; Kim, Y.S. Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J. Sci. Food Agric. 2010, 90, 1926–1935. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kwak, H.S.; Jung, H.Y.; Kim, S.S. Microbial communities related to sensory attributes in Korean fermented soy bean paste (doenjang). Food Res. Int. 2016, 89 Pt 1, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.Y.; Jung, J.Y.; Lee, H.J.; Jeon, C.O. Functional characterization of bacterial communities responsible for fermentation of Doenjang: A traditional Korean fermented soybean paste. Front. Microbiol. 2016, 7, 827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, X.; Li, M.; Liu, Y.; Zhang, X.; Zheng, Y. Microbial diversity and function of soybean paste in East Asia: What we know and what we don’t. Curr. Opin. Food Sci. 2021, 37, 145–152. [Google Scholar] [CrossRef]





| Groups | Times (h) | Quality Properties (%, Except for pH) | Viable Microbial Counts (log CFU/g) | ||||
|---|---|---|---|---|---|---|---|
| Moisture | pH | Titratable Acidity | Amino-Type Nitrogen | Aerobic Bacteria | Mold | ||
| A | 0 | 60.12 ± 1.90 a | 6.52 ± 0.01 e | 0.38 ± 0.00 ef | 180.0 ± 11.1 d | 2.30 ± 0.00 bc | 0.00 ± 0.00 d |
| 24 | 59.21 ± 2.97 a | 6.57 ± 0.00 b | 0.42 ± 0.00 d | 226.0 ± 11.0 bc | 7.93 ± 0.06 a | 0.00 ± 0.00 d | |
| B | 0 | 60.03 ± 2.31 a | 6.62 ± 0.01 a | 0.32 ± 0.01 g | 260.0 ± 11.1 b | 0.00 ± 0.00 c | 5.46 ± 0.17 b |
| 24 | 59.96 ± 1.66 a | 6.28 ± 0.00h | 0.78 ± 0.00 b | 776.0 ± 11.1 a | 8.41 ± 0.26 a | 6.45 ± 0.16 a | |
| C | 0 | 58.18 ± 0.71 a | 6.54 ± 0.00 d | 0.37 ± 0.00 f | 227.0 ± 11.1 bc | 2.15 ± 3.04 bc | 4.67 ± 0.02 c |
| 24 | 58.15 ± 1.35 a | 6.32 ± 0.00 g | 0.95 ± 0.00 a | 776.0 ± 11.1 a | 9.40 ± 0.02 a | 6.44 ± 0.04 a | |
| D | 0 | 58.66 ± 0.32 a | 6.46 ± 0.00 f | 0.40 ± 0.01 e | 196.0 ± 11.1 cd | 5.77 ± 0.38 ab | 5.62 ± 0.09 b |
| 24 | 57.66 ± 0.29 a | 6.55 ± 0.00 c | 0.68 ± 0.01 c | 172.0 ± 0.0 d | 9.82 ± 0.05 a | 4.31 ± 0.08 c | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, N.-Y.; Jang, Y.-J.; Gwon, H.-M.; Jeong, W.-S.; Yeo, S.-H.; Kim, S.-Y. Comparative Evaluation of Quality and Metabolite Profiles in Meju Using Starter Cultures of Bacillus velezensis and Aspergillus oryzae. Foods 2022, 11, 68. https://doi.org/10.3390/foods11010068
Gil N-Y, Jang Y-J, Gwon H-M, Jeong W-S, Yeo S-H, Kim S-Y. Comparative Evaluation of Quality and Metabolite Profiles in Meju Using Starter Cultures of Bacillus velezensis and Aspergillus oryzae. Foods. 2022; 11(1):68. https://doi.org/10.3390/foods11010068
Chicago/Turabian StyleGil, Na-Young, Ye-Ji Jang, Hee-Min Gwon, Woo-Soo Jeong, Soo-Hwan Yeo, and So-Young Kim. 2022. "Comparative Evaluation of Quality and Metabolite Profiles in Meju Using Starter Cultures of Bacillus velezensis and Aspergillus oryzae" Foods 11, no. 1: 68. https://doi.org/10.3390/foods11010068