1. Introduction
Peas (
Pisum sativum L.) are increasingly used due to their sustainable production [
1], economic benefits [
2], high protein content (15–30%), and alleged low allergenicity. They belong to the legume family (Fabaceae) and their proteins are classified as salt-, water-, and ethanol soluble, corresponding to globulins, albumins, and prolamins, respectively [
3].
Peas are not on the list of main allergens and do not need to be declared as allergenic in food products; however, two allergenic protein fractions from the storage proteins have been identified [
4] and are recognized by the International Committee of Allergen Nomenclature as main pea globulin allergens. The allergen Pis s1 correspond to the mature vicilin (47–50 kDa) as well as to one of vicilin’s proteolytic fractions (32 kDa). The Pis s2 correspond to convicilin (67–70 kDa). The ability of allergen proteins (antigen) to cause an immune response (immunogenicity) depends on different factors, such as the antigen dose, exposure, and host genetic background [
5], and thus, their ability to cause allergic reactions [
6]. Moreover, pea allergens have shown homology between epitopes (recognition sites) from other legume allergens [
7] and serological cross-reactivity has been proved [
4,
8,
9]. Different methods to modify food allergens and their impact on food allergenicity have been reviewed [
10].
Enzymatic hydrolysis is one of the most common methods used for this purpose and has been proven effective in allergen degradation of different legumes [
11,
12,
13]. Modification of pea allergens by enzymatic treatment has been studied to a lesser extent. Pea protein isolate (PPI) treated with trypsin [
14], Alcalase [
15], flavourzyme, papain, and pepsin [
16] have shown a reduced immunogenicity by means of ELISA methods. Frączek, Kostyra [
14] found that a higher degree of hydrolysis resulted in a higher reduction in immunogenic potential. Moreover, changes in the molecular weight distribution of proteins are also known to affect functional and sensory properties. Partial hydrolysis was shown to increase protein solubility and emulsifying capacity; however, further hydrolysis reduced both [
17,
18,
19]. Depending on the composition, the low molecular weight peptides formed during enzymatic hydrolysis can promote a bitter taste. The mechanism is not yet fully understood but mainly hydrophobic amino acid residues appear responsible [
20].
For debittering of protein hydrolysates, fermentation has been widely studied [
21,
22,
23,
24,
25]. Lactic acid bacteria reduced the bitterness of hydrolysates by releasing aminopeptidases cleaving hydrophobic amino acid residues [
23]. There are several studies focusing on changes in the aroma profile of fermented pea, pea proteins, and pea products [
26,
27,
28]; however, to our knowledge, there are no studies focusing on the debittering of pea protein hydrolysates by lactic fermentation.
The effects of fermentation on the functional properties have been studied for different legumes [
21,
29,
30], and, to a lesser extent, for peas [
31,
32,
33]. Moreover and to our knowledge, only one study has investigated the effects of fermentation on the antigenicity of pea flour [
34].
The combination of enzymatic hydrolysis and microbial fermentation seems very promising for the production of low-allergenic and tasty functional food ingredients. A combination has been investigated for soy [
22] and lupin protein isolate [
35,
36], but not yet for pea. For this reason, this study aimed to investigate the effects of combining enzymatic hydrolysis and fermentation on allergenic proteins (Pis s1 and Pis s2), as measured by SDS-PAGE and the ability of polyclonal sera to recognize antigens, functional properties and on the debittering and characteristic off-flavors of pea proteins. According to previous findings, papain, Esperase
®, trypsin and
Lactobacillus plantarum were selected for enzymatic hydrolysis [
18] and fermentation [
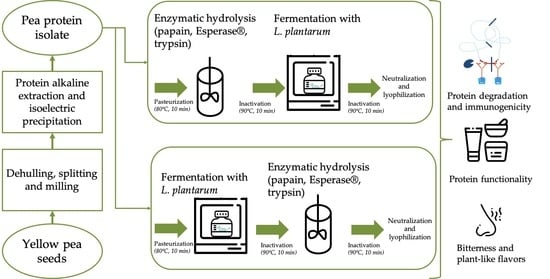
33], respectively. The specificity of an enzyme is determined by the arrangement of amino acids within the active site and the structure of the substrates. The acidification during fermentation could cause protein aggregation hiding protein parts from binding with the enzyme active site. Therefore, two sequences of the reactions, enzymatic treatment and fermentation, were investigated as the order of the method combination might be relevant for changes in the molecular weight distribution of the hydrolysates, functional properties and taste. Moreover, the treatments and the order of the method combination might also change the epitope binding sites and thus, the immunogenicity of pea allergenic proteins.
3. Results and Discussion
3.1. Microbial Growth
L. plantarum requires tryptophan, arginine, glutamate and branched-chain amino acids (isoleucine, leucine, valine) for growth [
46]. Besides of tryptophan, PPI is a good source of all the required amino acids; thus,
L. plantarum was able to grow both, in the PPI dispersion and hydrolyzed PPI (
Table 3). However, fermentation of PPI hydrolysates resulted in significantly higher viable cell counts compared to the fermented PPI. This could be due to some release of amino acids and peptides during hydrolysis, which provide a readily available source of nutrients for
L. plantarum growth. The hydrolysates showed slight differences in CFU after fermentation, with P_HyF showing the highest value of 9.53 Log CFU/mL followed by E_HyF and T_HyF with 9.30 Log CFU/mL and 9.17 Log CFU/mL, respectively. The pH was measured after 24 h of fermentation and was similar for all fermented samples (pH 4.5 ± 0.2). A recent study showed that lactic fermentation of hydrolyzed lupin protein isolate resulted in similar pH values regardless of the enzyme used [
35].
3.2. Chemical Composition
The untreated PPI showed a protein content of 84.7 ± 0.1% (
Table 4). The average protein content of PPI hydrolysates (83.4 ± 1.4%) was significantly higher compared to fermented PPI (79.5 ± 0.3%) and to the average of the samples produced by the combination of both treatments (76.6 ± 1.3%). The differences in protein contents might be due partial metabolism of the proteins and increase in organic acids such as lactic acid and, in lesser extent, acetic acid [
46]. In addition, the ash content could be attributed to the addition of inorganic acid (hydrochloric acid) and sodium hydroxide to adjust the pH for each sample conditions.
3.3. Proteolysis of PPI
The SDS-PAGE and gel filtration were performed to observe the effects of the different treatments on the pea proteins. The molecular weight distribution of the untreated PPI and treated samples is shown in
Figure 1 and the positions of the main allergens are marked. The untreated PPI showed protein fractions between 97.5 and 6.5 kDa. The fermented PPI did not show major changes in the electrophoretic pattern as previously shown by García Arteaga, Leffler [
33] for six lactic fermentations. The enzymatic hydrolysis facilitated significant changes in the molecular weight distribution of the respective samples with an increase in smaller peptides. This was observed in the samples that were only enzymatically hydrolyzed as well as in the samples with combined methods. The protein pattern of the sample treated with papain (P_Hy) only showed bands smaller than 40 kDa—with the exception of one band around 69.1 kDa. This band was degraded by the subsequent fermentation (P_HyF) and only bands smaller than 27 kDa were found. The proteolysis with Esperase
® (E_Hy) and trypsin (T_Hy) resulted in protein fractions below 40 kDa and 34 kDa, respectively. Fermentation of these hydrolysates (E_HyF and T_HyF) did not change the molecular weight distribution, while hydrolysis after fermentation (FdH) resulted in further protein degradation with protein fractions smaller than 27 kDa.
Fermentation alone did not lead to large changes in the molecular weight distribution of the respective samples, probably due to the inability of
L. plantarum to metabolize large polypeptides [
47]. Enzymatic hydrolysis enhanced the degradation of large polypeptides into smaller peptides that can be easily metabolized by
L. plantarum [
47]. Furthermore,
L. plantarum activates peptidases with higher specificity for hydrophobic dipeptides [
46]. Proteolysis is known to release hydrophobic amino acids and peptides, which then can be digested by the lactic acid bacteria.
The samples that were first fermented and then enzymatically hydrolyzed showed protein fractions below 26 kDa. One explanation might be that due to the low pH, partial acid hydrolysis occurred during fermentation, and the enzymes then further broke down these hydrolyzed fractions.
3.3.1. Effect of Combined Methods on Pea Protein Allergens
A protein band at 63–80 kDa [
4,
18,
48,
49] could represent the Pis s2 allergen. In the present study, a protein band around 70.9 ± 0.9 kDa was found in the untreated PPI and with reduced intensity in the fermented PPI; this fraction could correspond to the Pis s2. The reduction could be due to a reduction in protein solubility (as explained later in
Section 3.5.1) rather than to a proteolytic effect of fermentation with
L. plantarum. Furthermore, P_Hy also showed this allergen fraction with a slightly lower intensity than the untreated PPI. This could explain that papain alone was not able to cleave this fraction.
Protein bands found around 50.1 ± 0.8 kDa and 31.91 ± 0.5 kDa could correspond to the Pis s1 of the mature vicilin (αβγ) and its proteolytic fraction (αβ), respectively. The Pis s1 αβγ was present in the untreated PPI and with less intensity in the fermented PPI. Its proteolytic fraction was present in the untreated PPI, fermented PPI, E_Hy, T_Hy, E_HyF, and T_HyF.
3.3.2. Effect of Combined Methods on the Degree of Hydrolysis
Both trypsin and Esperase
® are serine endoproteases, with trypsin having specificity for basic residues, such as lysine and arginine derivatives [
50] and Esperase
® having a broader specificity, such as for both hydrophobic and hydrophilic residues [
51]. The latter might explain the higher DH of all Esperase
® treated samples (
Table 5). Papain cleaves peptide bonds C-terminal of glycine and cysteine residues among others [
52]. Glycine and cysteine residues might interfere with the OPA agent giving unstable and weak signals [
40,
53]. This effect might have been the reason why papain treated samples showed lower DH compared to other hydrolyzed samples even when the electrophoretic results showed significant changes.
Furthermore, the combination of fermentation after enzymatic hydrolysis significantly increased the DH value compared to the untreated PPI, the fermented PPI and the enzymatic treated sample. This could be related to the aforementioned ability of L. plantarum to take up the smaller peptides released after enzymatic hydrolysis. However, P_FdH and E_FdH did not show significant differences to P_Hy and E_Hy samples, respectively. In the case of P_FdH, this could be due to the higher exposure of cysteine residues interfering with the measurement; in the case of E_FdH, this could be due to protein agglomeration promoted by fermentation, which hides the cleavage site for Esperase®.
3.4. Reaction of Polyclonal Antibodies with PPI
The soluble proteins from all samples were separated by gel filtration and individual fractions analyzed by ELISA using three individual polyclonal rabbit sera raised against PPI. The results from all treated samples showed a compelling degradation towards lower molecular weight proteins (
Figure 2A). ELISA analyses of the total protein (
Figure 2B) and individual fractions gave a reduced immunogenicity for all samples. In particular, trypsin-treated samples showed a reduced antibody reactivity to background levels (
Figure 3A,B). Since the three polyclonal sera used for the ELISA showed different binding profiles for the individual proteins, it can be concluded that the soluble proteins are no longer recognized by the antibodies. The only exception are the eluted fractions containing higher molecular weight proteins, which are certainly resistant to the treatment applied. The ELISA results for the total protein showed that also the overall signal is significantly reduced in those preparations with the highest degradation. Therefore, the fraction of high molecular weight immunogenic proteins may be lower than suggested by the ELISA values of the high molecular weight fractions.
These SDS-PAGE results in combination with the results from the gel filtration and ELISA show that the combination of enzymatic hydrolysis and fermentation degrades pea proteins to a higher degree. Reduced reactivity with the antibody sera could imply reduction of the allergic potential of pea protein preparations. Fermentation followed by enzymatic hydrolysis was particularly successful, as it seemed to degrade all major potential pea allergens. However, the reduction in allergenicity needs to be confirmed by further immunological studies, such as prick tests.
3.5. Functional Properties
Changes in the molecular weight distribution of proteins cause changes in the exposed hydrophobic and ionizable groups as well as in the ability of the proteins to aggregate, which can influence the functional properties [
54]. Therefore, the effect of protein degradation on functional properties were studied in detail.
3.5.1. Protein Solubility
The results of the protein solubility analyses are shown in
Table 6; these results correlate strongly with the DH values. At acidic pH (pH 4.5), the untreated and fermented PPI were significantly different from all other samples. Samples treated with Esperase
® showed the highest protein solubility of up to 66%, whereas the protein solubility of papain and trypsin treated samples was also significantly increased. The fermentation followed by enzymatic hydrolysis was most effective in increasing solubility at acidic pH.
At neutral pH, the fermented PPI showed significant lower protein solubility compared to the untreated PPI. The PPI showed similar protein solubility to P_Hy and P_HyF, whereas the P_FdH was significantly different. The papain and trypsin treated samples showed the highest protein solubility when the fermentation step was followed by enzymatic hydrolysis. Among the samples that were only hydrolyzed or were hydrolyzed and then fermented, a significant difference in protein solubility could not be measured. However, the samples treated with Esperase® were significantly different from each other and from the untreated PPI. The lower solubility of E_FdH compared to E_HyF could be explained by an increase in insoluble aggregates due to acid denaturation during fermentation, which hinders the Esperase® activity to cleave on specific protein sites.
Other studies have shown negative or no effect of fermentation on the protein solubility. This has been attributed to changes in the protein surface, surface charge and the LAB cell surface, which might promote hydrophobic interactions [
33,
47,
55]. Thus, the improvement in the protein solubility of treated samples is certainly due to enzymatic hydrolysis.
3.5.2. Emulsifying Capacity
Results from emulsifying capacity are shown in
Table 6. The untreated PPI showed the highest emulsifying capacity with 725 mL/g, followed by T_FdH with 700 mL/g. In contrast, the fermented PPI and the E_HyF showed the lowest emulsifying capacity with 310 mL/g and 300 mL/g, respectively. The difference among the results of the treated samples could be due to different changes in protein conformation, peptide release, and their interactions with other components such as microbial cells, which could reduce the amphiphilic character of the proteins [
56]. Moreover, the ratio albumin/legumin/vicilin, the presence of polar lipids and partial denaturation have also been shown to affect emulsifying capacity [
57,
58,
59,
60]. Although all treated samples had lower emulsifying capacities than the untreated PPI, the emulsifying capacity of the treated samples is still in a good range to be used as food ingredient. A high DH is known to impair emulsifying capacities [
54], and although there was no correlation between the DH value and the emulsifying capacity, the sample with the highest DH (E_HyF) showed the lowest emulsifying capacity.
3.5.3. Foaming Capacity
A foam is a dispersion of air in water. The effect of proteins in foam formation is similar to the one in forming emulsions. Their amphiphilic character allows proteins to interact with the hydrophobic and hydrophilic fractions of air and water, respectively, during whipping, reducing surface tension. Similarly, the foaming capacity depends on different factors such as protein fractions ratio, pH of the solutions, and lipid content [
61,
62].
The foaming capacities of untreated and fermented PPI were not significantly different with 840% and 807%, respectively. On the other hand, all other treated samples showed a significantly improved foaming capacity, with the highest foaming capacity found in the fermented and subsequently hydrolyzed samples. Of the treated samples, the trypsin samples showed the highest capacities.
3.6. Sensory Analysis
Although products containing pea proteins are increasing, the characteristic pea off-flavors remain a major challenge. In addition to naturally occurring off-flavors, PPI treatment can lead to changes in the flavor and taste profile. It is known that enzymatic hydrolysis can increase the bitterness of protein preparations from legumes, whereas fermentation of legumes promotes the degradation and formation of aroma compounds.
The bitterness of the untreated PPI compared to those treated with papain or trypsin was not significantly different (
Figure 4). However, the bitterness ranking was the highest after treatment with Esperase
® and was significantly higher than that of the untreated PPI. Although fermentation enhanced further hydrolysis (
Section 3.3), the peptidases from
L. plantarum may not be sufficient to completely cleave hydrophobic residues.
As expected, the untreated PPI was ranked highest for plant-like off-flavor, while this attribute was significantly reduced for all combined samples. The samples fermented prior to enzymatic hydrolysis showed the strongest reduction of the plant-like off-flavor, where T_FdH received the lowest rank.