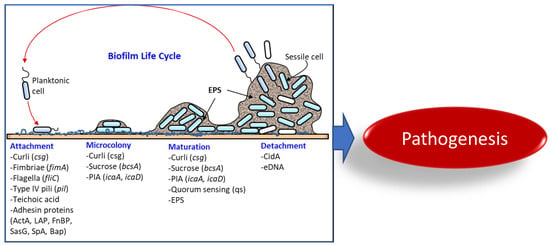
Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety
Abstract
:1. Introduction
Bacterial Biofilms and Food Safety Concerns
2. Bacterial Virulence Factors that Contribute to Biofilm Formation and Pathogenesis

2.1. Listeria monocytogenes Cell Surface-Associated Adhesion Molecules Are Involved in Host Cell Interaction and Biofilm Formation
2.1.1. Role of Sigma B, PrfA, and ActA on Biofilm Formation and Pathogenesis
2.1.2. Listeria Teichoic Acid
2.1.3. Listeria Adhesion Protein
2.2. Staphylococcus aureus Surface Proteins Play Important Roles in Biofilm Formation and Pathogenesis
2.2.1. Polysaccharide Intercellular Adhesin (PIA)
2.2.2. S. aureus Protein A
2.2.3. Biofilm-Associated Protein (Bap)
2.2.4. Fibronectin-Binding Proteins (FnBP)
2.2.5. S. aureus Surface Protein G (SasG)
2.2.6. Staphylococcal Teichoic Acid
2.2.7. Miscellaneous Factors
2.3. Escherichia coli Curli Fimbriae, and Cellulose Are Major Biofilm-Forming Factors
2.3.1. E. coli Curli
2.3.2. Cellulose
2.3.3. Aggregative Adherence Fimbriae (AAF)
| Bacteria | Factors | Function | Refs | |
|---|---|---|---|---|
| Biofilm Formation | Pathogenicity | |||
| Listeria monocytogenes | ActA (actin polymerization protein) | Bacterial sedimentation and aggregation | Rearrange host cytoskeletal structure and promote the cell-to-cell spread | [75] |
| LAP (listeria adhesion protein) | Expression in recombinant Lactobacillus enhanced biofilm formation | Epithelial adhesion and translocation through the epithelial barrier | [50,51] | |
| PrfA (protein regulatory factor) | Regulate the expression of ActA that is necessary for biofilm formation | Regulatory protein that regulates the synthesis of multiple virulence factors | [71] | |
| WTA (wall teichoic acid) | Maintain cell wall (peptidoglycan) architecture and participate in biofilm formation | Induce inflammatory response | [77] | |
| Staphylococcus aureus | Bap (biofilm-associated protein) | Adhesion to inert surfaces and intercellular adhesion in the development of biofilm formation | Establish persistent infection on a mouse infection model | [101,103] |
| Protein A | Cell-to-cell adhesion in biofilm development; a major proteinaceous component in S. aureus biofilms | Help S. aureus to evade immune system in vivo | [99,100] | |
| PIA (polysaccharide intercellular adhesin) | Cell-to-cell binding in biofilm formation | Establish persistent in vivo infection | [91,93] | |
| Teichoic acid | Maintain cell wall (peptidoglycan) architecture and participate in biofilm formation | Induce inflammatory response | [76,77,111] | |
| FnBP (fibronectin-binding proteins) | Cell-to-cell adhesion through low-affinity homophilic interaction between neighboring cells | Promote bacterial attachment to host fibronectin for adhesion and colonization | [105,107] | |
| SasG (S. aureus surface protein G) | Zinc activated SasG-mediated biofilm formation | Adhesion to epithelial cells | [89,109] | |
| Salmonella enterica | Fimbria (SEF17) | Cell-to-cell interaction in biofilm formation | Bind to human fibronectin and facilitate cell invasion | [144,145] |
| Bap (biofilm-associated protein) | Bap and curli can help form strong biofilms in both biotic and abiotic surface | Colonization, intestinal persistence, invasion to liver and spleen and lethality in mice | [126] | |
| CsgD, BcsA | Curli and cellulose synthesis | Colonization, biofilm formation and vertical transmission to egg | [146] | |
| Escherichia coli | Curli made with CsgA and CsgB | Adherence to abiotic surfaces | Adhere to epithelial cells when over expressed | [129,135] |
| Fim (fimbriae) | Biofilm formation on polystyrol | Adhesion to epithelial cell lines | [136] | |
| Enteroaggregative E. coli (EAEC) | Aggregative adherence fimbriae (AAF) | Mediate biofilm formation on abiotic surfaces | Bind to MUC1 on epithelial cells | [142,143] |
| Pseudomonas aeruginosa | PqsR | A key component of Pseudomonas quinolone signal system | Regulate the production of virulence factors, pyocyanin and hydrogen cyanide | [147] |
| Flagellum | Swimming motility and biofilm formation | Flagella is an important virulence factor. The flagellum-deficient strain showed less invasion in the mouse burn wound model and less colonization in the murine intestine | [148,149] | |
| Type IV pili | Twitching motility, and adhesion to abiotic surfaces | Adhesion to eukaryotic cells and pathogenesis | [150] | |
2.4. Salmonella enterica Curli Fimbriae and Bap Play Important Roles in Biofilm Formation and Pathogenesis
2.4.1. Salmonella Curli
2.4.2. Salmonella Biofilm-Associated Protein (Bap)
2.5. Pseudomonas aeruginosa, an Opportunistic Pathogen that Forms a Strong Biofilm
2.5.1. Pseudomonas Flagella and Pili Aid in the Initial Attachment
2.5.2. Pseudomonas Microcolony Formation and Biofilm Maturation Are Regulated by a Quorum-Sensing Network
3. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015, 9, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [Green Version]
- Yaron, S.; Romling, U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 2014, 7, 496–516. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600. [Google Scholar] [CrossRef]
- Pinto, M.; Langer, T.M.; Hüffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef] [Green Version]
- CDC, Multistate Outbreak of Listeriosis Linked to Whole Cantaloupes from Jensen Farms, Colorado (FINAL UPDATE). Available online: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html (accessed on 27 August 2021).
- Fu, Y.; Deering, A.J.; Bhunia, A.K.; Yao, Y. Pathogen biofilm formation on cantaloupe surface and its impact on the antibacterial effect of lauroyl arginate ethyl. Food Microbiol. 2017, 64, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Martins, K.B.; Ferreira, A.M.; Pereira, V.C.; Pinheiro, L.; Oliveira, A.D.; Cunha, M.D.L.R.D.S.D. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus saprophyticus isolated from patients with urinary tract infections. Front. Microbiol. 2019, 10, 40. [Google Scholar] [CrossRef]
- Kubota, H.; Senda, S.; Tokuda, H.; Uchiyama, H.; Nomura, N. Stress resistance of biofilm and planktonic Lactobacillus plantarum subsp. plantarum JCM 1149. Food Microbiol. 2009, 26, 592–597. [Google Scholar] [CrossRef] [Green Version]
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. Npj Biofilm. Microbiome 2016, 2, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, S.H.; Tse, E.C.M.; Yates, M.D.; Otero, F.J.; Trammell, S.A.; Stemp, E.D.A.; Barton, J.K.; Tender, L.M.; Newman, D.K. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 2020, 182, 919–932.e19. [Google Scholar] [CrossRef]
- Ray, B.; Bhunia, A. Microbial attachements and biofilm formation. In Fundamental Food Microbiology, 5th ed.; Ray, B., Bhunia, A., Eds.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar] [CrossRef]
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes biofilms in the food industry: Is the current hygiene program sufficient to combat the persistence of the pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef] [PubMed]
- Shirron, N.; Kisluk, G.; Zelikovich, Y.; Eivin, I.; Shimoni, E.; Yaron, S. A comparative study assaying commonly used sanitizers for antimicrobial activity against indicator bacteria and a Salmonella Typhimurium strain on fresh produce. J. Food Prot. 2009, 72, 2413–2417. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Doepfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef] [Green Version]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef]
- Kim, S.O.; Kim, S.S. Recent (2011–2017) foodborne outbreak cases in the Republic of Korea compared to the United States: A review. Food Sci. Biotechnol. 2021, 30, 185–194. [Google Scholar] [CrossRef]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Ponniah, J.; Robin, T.; Paie, M.S.; Radu, S.; Ghazali, F.M.; Kqueen, C.Y.; Nishibuchi, M.; Nakaguchi, Y.; Malakar, P.K. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Conrol 2010, 21, 774–778. [Google Scholar] [CrossRef]
- Wu, S.; Wu, Q.; Zhang, J.; Chen, M.; Hu, H. Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS ONE 2015, 10, e0136682. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef]
- Reis-Teixeira, F.B.D.; Alves, V.F.; Martinis, E.C.P.D. Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Brazilian J. Microbiol. 2017, 48, 587–591. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F.; Kathariou, S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 2006, 72, 7711–7717. [Google Scholar] [CrossRef] [Green Version]
- Langsrud, S.; Moen, B.; Møretrø, T.; Løype, M.; Heir, E. Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J. Appl. Microbiol. 2016, 120, 366–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Xie, J.; Soteyome, T.; Peters, B.M.; Shirtliff, M.E.; Liu, J.; Harro, J.M. Polymicrobial interaction and biofilms between Staphylococcus aureus and Pseudomonas aeruginosa: An underestimated concern in food safety. Curr. Opin. Food Sci. 2019, 26, 57–64. [Google Scholar] [CrossRef]
- Dalton, C.B.; Austin, C.C.; Sobel, J.; Hayes, P.S.; Bibb, W.F.; Graves, L.M.; Swaminathan, B.; Proctor, M.E.; Griffin, P.M. An Outbreak of Gastroenteritis and Fever Due to Listeria monocytogenes in Milk. N. Engl. J. Med. 1997, 336, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Pouillot, R.; Klontz, K.C.; Chen, Y.; Burall, L.S.; Macarisin, D.; Doyle, M.; Bally, K.M.; Strain, E.; Datta, A.R.; Hammack, T.S. Infectious dose of Listeria monocytogenes in outbreak linked to ice cream, United States, 2015. Emerg. Infect. Dis. 2016, 22, 2113–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhunia, A.K. Staphylococcus aureus. In Foodborne Microbial Pathogens: Mechanisms and Pathogenesis; Bhunia, A.K., Ed.; Springer: New York, NY, USA, 2018; pp. 181–192. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kothary, M.H.; Babu, U.S. Infective dose of foodborne pathogens in volunteers: A review. J. Food Safety 2001, 21, 49–68. [Google Scholar] [CrossRef]
- van den Brom, R.; de Jong, A.; van Engelen, E.; Heuvelink, A.; Vellema, P. Zoonotic risks of pathogens from sheep and their milk borne transmission. Small Rumin. Res. 2020, 189, 106123. [Google Scholar] [CrossRef] [PubMed]
- Roser, D.J.; van den Akker, B.; Boase, S.; Haas, C.N.; Ashbolt, N.J.; Rice, S.A. Pseudomonas aeruginosa dose response and bathing water infection. Epidemiol. Infect. 2014, 142, 449–462. [Google Scholar] [CrossRef]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, L.; Han, J.; Dong, P.; Luo, X.; Zhang, Y.; Zhu, L. Inhibition of Biofilm Formation and Related Gene Expression of Listeria monocytogenes in Response to Four Natural Antimicrobial Compounds and Sodium Hypochlorite. Front. Microbiol. 2021, 11, 3523. [Google Scholar] [CrossRef]
- Almeida, F.A.D.; Vargas, E.L.G.; Carneiro, D.G.; Pinto, U.M.; Vanetti, M.C.D. Virtual screening of plant compounds and nonsteroidal anti-inflammatory drugs for inhibition of quorum sensing and biofilm formation in Salmonella. Microb. Pathog. 2018, 121, 369–388. [Google Scholar] [CrossRef]
- Aswathanarayan, J.B.; Vittal, R.R. Inhibition of biofilm formation and quorum sensing mediated phenotypes by berberine in Pseudomonas aeruginosa and Salmonella Typhimurium. RSC Adv. 2018, 8, 36133–36141. [Google Scholar] [CrossRef] [Green Version]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.K.; Zhao, A.; Wang, A.; Brown, Z.Z.; Muir, T.W.; Stone, H.A.; Bassler, B.L. Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2017, 2, 17080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes-Luz, L.; Mendonça, M.; Bernardes Fogaça, M.; Kipnis, A.; Bhunia, A.K.; Bührer-Sékula, S. Listeria monocytogenes: Review of pathogenesis and virulence determinants-targeted immunological assays. Crit. Rev. Microbiol. 2021, 47, 647–666. [Google Scholar] [CrossRef]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Drolia, R.; Bhunia, A.K. Crossing the intestinal barrier via Listeria adhesion protein and internalin A. Trends Microbiol. 2019, 27, 408–425. [Google Scholar] [CrossRef]
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp60. Infect. Immun. 2010, 78, 5062–5073. [Google Scholar] [CrossRef] [Green Version]
- Jagadeesan, B.; Koo, O.K.; Kim, K.P.; Burkholder, K.M.; Mishra, K.K.; Aroonnual, A.; Bhunia, A.K. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 2010, 156, 2782–2795. [Google Scholar] [CrossRef] [PubMed]
- Drolia, R.; Tenguria, S.; Durkes, A.C.; Turner, J.R.; Bhunia, A.K. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 2018, 23, 470–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drolia, R.; Amalaradjou, M.A.R.; Ryan, V.; Tenguria, S.; Liu, D.; Bai, X.; Xu, L.; Singh, A.K.; Cox, A.D.; Bernal-Crespo, V.; et al. Receptor-targeted engineered probiotics mitigate lethal Listeria infection. Nat. Commun. 2020, 11, 6344. [Google Scholar] [CrossRef]
- Nikitas, G.; Deschamps, C.; Disson, O.; Niault, T.; Cossart, P.; Lecuit, M. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 2011, 208, 2263–2277. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Fevre, C.; Lavina, M.; Disson, O.; Lecuit, M. Live imaging reveals Listeria hijacking of E-cadherin recycling as it crosses the intestinal barrier. Curr. Biol. 2021, 31, 1037–1047.e4. [Google Scholar] [CrossRef]
- Gilmartin, N.; Gião, M.S.; Keevil, C.W.; O’Kennedy, R. Differential internalin A levels in biofilms of Listeria monocytogenes grown on different surfaces and nutrient conditions. Int. J. Food Microbiol. 2016, 219, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Gao, Z.; Zhang, Z.; Pan, L.; Zhang, Y. Roles of M cells in infection and mucosal vaccines. Hum. Vacc. Immunother. 2014, 10, 3544–3551. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Nagai, T.; Hayashi, T.; Baba, Y.; Nagai, S.; Koyasu, S. Listerial invasion protein internalin B promotes entry into ileal Peyer’s patches in vivo. Microbiol. Immunol. 2011, 55, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Pentecost, M.; Kumaran, J.; Ghosh, P.; Amieva, M.R. Listeria monocytogenes Internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 2010, 6, e1000900. [Google Scholar] [CrossRef] [PubMed]
- Gessain, G.; Tsai, Y.-H.; Travier, L.; Bonazzi, M.; Grayo, S.; Cossart, P.; Charlier, C.; Disson, O.; Lecuit, M. PI3-kinase activation is critical for host barrier permissiveness to Listeria monocytogenes. J. Exp. Med. 2015, 212, 165–183. [Google Scholar] [CrossRef] [Green Version]
- Osborne, S.E.; Brumell, J.H. Listeriolysin O: From bazooka to Swiss army knife. Phil. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160222. [Google Scholar] [CrossRef] [Green Version]
- Mulvihill, E.; van Pee, K.; Mari, S.A.; Müller, D.J.; Yildiz, Ö. Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 2015, 15, 6965–6973. [Google Scholar] [CrossRef]
- Kortebi, M.; Milohanic, E.; Mitchell, G.; Péchoux, C.; Prevost, M.-C.; Cossart, P.; Bierne, H. Listeria monocytogenes switches from dissemination to persistence by adopting a vacuolar lifestyle in epithelial cells. PLoS Pathog. 2017, 13, e1006734. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, D.A.; Chakraborty, T.; Goebel, W.; Cossart, P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 1992, 60, 1263–1267. [Google Scholar] [CrossRef] [Green Version]
- Gouin, E.; Adib-Conquy, M.; Balestrino, D.; Nahori, M.-A.; Villiers, V.; Colland, F.; Dramsi, S.; Dussurget, O.; Cossart, P. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the I kappa B kinase subunit IKK alpha. Proc. Natl. Acad. Sci. USA 2010, 107, 17333–17338. [Google Scholar] [CrossRef] [Green Version]
- Kocks, C.; Gouin, E.; Tabouret, M.; Berche, P.; Ohayon, H.; Cossart, P. L. monocytogenes -induced actin assembly requires the actA gene product, a surface protein. Cell 1992, 68, 521–531. [Google Scholar] [CrossRef]
- Cheng, M.I.; Chen, C.; Engström, P.; Portnoy, D.A.; Mitchell, G. Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell. Microbiol. 2018, 20. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, G.; Ge, L.; Huang, Q.; Chen, C.; Kianian, S.; Roberts, M.F.; Schekman, R.; Portnoy, D.A. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 2015, 83, 2175–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tattoli, I.; Sorbara, M.T.; Yang, C.; Tooze, S.A.; Philpott, D.J.; Girardin, S.E. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J. 2013, 32, 3066–3078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vazquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Guariglia-Oropeza, V.; Wiedmann, M.; Boor, K.J. Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra-and intracellular environments. Microiol. Mol. Biol. Rev. 2019, 83, e00034-19. [Google Scholar] [CrossRef]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef]
- Lemon, K.P.; Freitag, N.E.; Kolter, R. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J. Bacteriol. 2011, 192, 3969–3976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorey, A.; Marinho, C.; Piveteau, P.; O’Byrne, C. Role and regulation of the stress activated sigma factor sigma B (σ(B)) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 2019, 106, 1–48. [Google Scholar] [CrossRef]
- Horn, N.; Bhunia, A.K. Food-Associated Stress Primes Foodborne Pathogens for the Gastrointestinal Phase of Infection. Front. Microbiol. 2018, 9, 1962. [Google Scholar] [CrossRef] [Green Version]
- van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travier, L.; Guadagnini, S.; Gouin, E.; Dufour, A.; Chenal-Francisque, V.; Cossart, P.; Olivo-Marin, J.-C.; Ghigo, J.-M.; Disson, O.; Lecuit, M. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013, 9, e1003131. [Google Scholar] [CrossRef] [Green Version]
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Liu, D.; Singh, A.K.; Drolia, R.; Bai, X.; Tenguria, S.; Bhunia, A.K. Tunicamycin mediated inhibition of wall teichoic acid affect Staphylococcus aureus and Listeria monocytogenes cell morphology, biofilm formation and virulence. Front. Microbiol. 2018, 9, 1352. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Liu, D.; Xu, L.; Tenguria, S.; Drolia, R.; Gallina, N.L.F.; Cox, A.D.; Koo, O.-K.; Bhunia, A.K. Biofilm-isolated Listeria monocytogenes exhibits reduced systemic dissemination at the early (12–24 h) stage of infection in a mouse model. Npj Biofilm. Microbiome 2021, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Liang, Y.; Chen, L.; Wang, W.; Wang, J.; Li, B.; Li, L.; Chen, D.; Xu, Z. Formation and development of Staphylococcus biofilm: With focus on food safety. J. Food Saftey 2017, 37, e12358. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef] [Green Version]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation, and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Bennett, S.D.; Walsh, K.A.; Gould, L.H. Foodborne disease outbreaks caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus—United States, 1998–2008. Clin. Infect. Dis. 2013, 57, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Pillsbury, A.; Chiew, M.; Bates, J.; Sheppeard, V. An outbreak of staphylococcal food poisoning in a commercially catered buffet. Commun. Dis. Intell. 2013, 37, E144–E148. [Google Scholar]
- Vitale, M.; Scatassa, M.L.; Cardamone, C.; Oliveri, G.; Piraino, C.; Alduina, R.; Napoli, C. Staphylococcal food poisoning case and molecular analysis of toxin genes in Staphylococcus aureus strains isolated from food in Sicily, Italy. Foodborne Pathog. Dis. 2015, 12, 21–23. [Google Scholar] [CrossRef]
- Di Ciccio, P.; Vergara, A.; Festino, A.R.; Paludi, D.; Zanardi, E.; Ghidini, S.; Ianieri, A. Biofilm formation by Staphylococcus aureus on food contact surfaces: Relationship with temperature and cell surface hydrophobicity. Food Control 2015, 50, 930–936. [Google Scholar] [CrossRef]
- Di Domenico, E.G.; Cavallo, I.; Capitanio, B.; Ascenzioni, F.; Pimpinelli, F.; Morrone, A.; Ensoli, F. Staphylococcus aureus and the Cutaneous Microbiota Biofilms in the Pathogenesis of Atopic Dermatitis. Microorganisms 2019, 7, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev. Anti-Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef] [Green Version]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef] [Green Version]
- Zapotoczna, M.; O’Neill, E.; O’Gara, J.P. Untangling the Diverse and Redundant Mechanisms of Staphylococcus aureus Biofilm Formation. PLoS Pathog. 2016, 12, e1005671. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1, 6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Gerke, C.; Kraft, A.; Süßmuth, R.; Schweitzer, O.; Götz, F. Characterization of theN-Acetylglucosaminyl transferase Activity Involved in the Biosynthesis of the Staphylococcus epidermidis Polysaccharide Intercellular Adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [Green Version]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenney, D.; Pouliot, K.L.; Wang, Y.; Murthy, V.; Ulrich, M.; Döring, G.; Lee, J.C.; Goldmann, D.A.; Pier, G.B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 1999, 284, 1523–1527. [Google Scholar] [CrossRef]
- Cramton, S.E.; Ulrich, M.; Götz, F.; Döring, G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69, 4079–4085. [Google Scholar] [CrossRef] [Green Version]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Bartscht, K.; Mack, D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 1999, 67, 2627–2632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francois, P.; Tu Quoc, P.H.; Bisognano, C.; Kelley, W.L.; Lew, D.P.; Schrenzel, J.; Cramton, S.E.; Götz, F.; Vaudaux, P. Lack of biofilm contribution to bacterial colonisation in an experimental model of foreign body infection by Staphylococcus aureus and Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 2003, 35, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Débarbouillé, M.; Penadés, J.R.; Lasa, I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef] [Green Version]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penadés, J.R.; Lasa, I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, S.D.; DeLeo, F.R. Staphylococcus aureus protein A promotes immune suppression. mBio 2013, 4, e00764-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, Í.; Penadés, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef] [Green Version]
- Trotonda, M.P.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 2005, 187, 5790–5798. [Google Scholar] [CrossRef] [Green Version]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.K.; Rao, T.S. Staphylococcus aureus biofilm removal by targeting biofilm-associated extracellular proteins. Indian J. Med. Res. 2017, 146, S1–S8. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, T.J. The remarkably multifunctional fibronectin binding proteins of Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Gries, C.M.; Biddle, T.; Bose, J.L.; Kielian, T.; Lo, D.D. Staphylococcus aureus fibronectin binding protein A mediates biofilm development and infection. Infect. Immun. 2020, 88, e00859-19. [Google Scholar] [CrossRef] [PubMed]
- Herman-Bausier, P.; El-Kirat-Chatel, S.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. mBio 2015, 6, e00413-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formosa-Dague, C.; Speziale, P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. USA 2016, 113, 410–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geoghegan, J.A.; Corrigan, R.M.; Gruszka, D.T.; Speziale, P.; O’Gara, J.P.; Potts, J.R.; Foster, T.J. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2010, 192, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Naclerio, G.A.; Onyedibe, K.I.; Sintim, H.O. Lipoteichoic Acid Biosynthesis Inhibitors as Potent Inhibitors of S. aureus and E. faecalis Growth and Biofilm Formation. Molecules 2020, 25, 2277. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Shi, L.; Wu, Y.; Yang, C.; Ma, Y.; Zhang, Q.-Z.; Huang, W.; Zhu, X.-Y.; Yan, Y.-J.; Wang, J.-X.; Zhu, T. Effect of nicotine on Staphylococcus aureus biofilm formation and virulence factors. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Guo, Y.; Zhan, Q.; Shang, Y.; Qu, D.; Yu, F. Subinhibitory Concentrations of Mupirocin Stimulate Staphylococcus aureus Biofilm Formation by Upregulating cidA. Antimicrob. Agents Chemother. 2020, 64, e01912-19. [Google Scholar] [CrossRef]
- Rice, K.C.; Firek, B.A.; Nelson, J.B.; Yang, S.-J.; Patton, T.G.; Bayles, K.W. The Staphylococcus aureus cidAB operon: Evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2003, 185, 2635–2643. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef] [Green Version]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef]
- Newell, D.G.; La Ragione, R.M. Enterohaemorrhagic and other Shiga toxin-producing Escherichia coli (STEC): Where are we now regarding diagnostics and control strategies? Transbound Emerg. Dis. 2018, 65, 49–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, E.; Cimdins, A.; Lüthje, P.; Brauner, A.; Sjöling, Å.; Landini, P.; Römling, U. “It’s a gut feeling”—Escherichia coli biofilm formation in the gastrointestinal tract environment. Crit. Rev. Microbiol. 2018, 44, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Currie, A.; Honish, L.; Cutler, J.; Locas, A.; Lavoie, M.-C.; Gaulin, C.; Galanis, E.; Tschetter, L.; Chui, L.; Taylor, M. Outbreak of Escherichia coli O157: H7 infections linked to mechanically tenderized beef and the largest beef recall in Canada, 2012. J. Food Prot. 2019, 82, 1532–1538. [Google Scholar] [CrossRef]
- de Oliveira Elias, S.; Noronha, T.B.; Tondo, E.C. Salmonella spp. and Escherichia coli O157:H7 prevalence and levels on lettuce: A systematic review and meta-analysis. Food Microbiol. 2019, 84, 103217. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.; Werber, D.; Cramer, J.P.; Askar, M.; Faber, M.; an der Heiden, M.; Bernard, H.; Fruth, A.; Prager, R.; Spode, A. Epidemic profile of Shiga-toxin–producing Escherichia coli O104: H4 outbreak in Germany. N. Engl. J. Med. 2011, 365, 1771–1780. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.; Doyle, M.P.; Phatak, S.C.; Millner, P.; Jiang, X. Persistence of enterohemorrhagic Escherichia coli O157: H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J. Food Prot. 2004, 67, 1365–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289. [Google Scholar] [CrossRef] [Green Version]
- Latasa, C.; Roux, A.; Toledo-Arana, A.; Ghigo, J.M.; Gamazo, C.; Penadés, J.R.; Lasa, I. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 2005, 58, 1322–1339. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.; Sharma, M.; Ravishakar, S. Effect of curli expression and hydrophobicity of Escherichia coli O157: H7 on attachment to fresh produce surfaces. J. Appl. Microbiol. 2011, 110, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.Q.; Botyanszki, Z.; Tay, P.K.R.; Joshi, N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Reichhardt, C.; Cegelski, L. The Congo red derivative FSB binds to curli amyloid fibers and specifically stains curliated E. coli. PLoS ONE 2018, 13, e0203226. [Google Scholar] [CrossRef]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [Green Version]
- Olsén, A.; Jonsson, A.; Normark, S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 1989, 338, 652–655. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Larsen, P.; Nielsen, J.L.; Otzen, D.; Nielsen, P.H. Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 2008, 74, 1517–1526. [Google Scholar] [CrossRef] [Green Version]
- Saldaña, Z.; Xicohtencatl-Cortes, J.; Avelino, F.; Phillips, A.D.; Kaper, J.B.; Puente, J.L.; Girón, J.A. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 2009, 11, 992–1006. [Google Scholar] [CrossRef] [Green Version]
- Elpers, L.; Hensel, M. Expression and functional characterization of various chaperon-usher fimbriae, curli fimbriae, and type 4 pili of enterohemorrhagic Escherichia coli O157:H7 Sakai. Front. Microbiol. 2020, 11, 378. [Google Scholar] [CrossRef]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Römling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef]
- Chirwa, N.T.; Herrington, M.B. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 2003, 149, 525–535. [Google Scholar] [CrossRef] [Green Version]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vial, P.A.; Robins-Browne, R.; Lior, H.; Prado, V.; Kaper, J.B.; Nataro, J.P.; Maneval, D.; Elsayed, A.-E.-D.; Levine, M.M. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 1988, 158, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czeczulin, J.R.; Whittam, T.S.; Henderson, I.R.; Navarro-Garcia, F.; Nataro, J.P. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 1999, 67, 2692–2699. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, J.; Hicks, S.; Dall’Agnol, M.; Phillips, A.D.; Nataro, J.P. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 2001, 41, 983–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boll, E.J.; Ayala-Lujan, J.; Szabady, R.L.; Louissaint, C.; Smith, R.Z.; Krogfelt, K.A.; Nataro, J.P.; Ruiz-Perez, F.; McCormick, B.A. Enteroaggregative Escherichia coli adherence fimbriae drive inflammatory cell recruitment via interactions with epithelial MUC1. mBio 2017, 8, e00717-17. [Google Scholar] [CrossRef] [Green Version]
- Austin, J.W.; Sanders, G.; Kay, W.W.; Collinson, S.K. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 1998, 162, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Collinson, S.; Doig, P.; Doran, J.; Clouthier, S.; Kay, W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Feng, Z.; Sun, H.; Zhang, R.; Qin, T.; Peng, D. Biofilm-Formation-Related Genes csgD and bcsA Promote the Vertical Transmission of Salmonella Enteritidis in Chicken. Front. Vet. Sci. 2020, 7, 625049. [Google Scholar] [CrossRef] [PubMed]
- Farrow, J.M.; Sund, Z.M.; Ellison, M.L.; Wade, D.S.; Coleman, J.P.; Pesci, E.C. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 2008, 190, 7043–7051. [Google Scholar] [CrossRef] [Green Version]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Pier, G.B.; Meluleni, G.; Goldberg, J.B. Clearance of Pseudomonas aeruginosa from the murine gastrointestinal tract is effectively mediated by O-antigen-specific circulating antibodies. Infect. Immun. 1995, 63, 2818–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Déziel, E.; Comeau, Y.; Villemur, R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 2001, 183, 1195–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Groisman, E.A. The origin and evolution of species differences in Escherichia coli and Salmonella typhimurium. In Molecular Ecology and Evolution: Approaches and Applications, 1994/01/01 ed.; Schierwater, B., Streit, B., Wagner, G.P., DeSalle, R., Eds.; Birkhäuser: Basel, Switzerland, 1994; Volume 69, pp. 479–493. [Google Scholar] [CrossRef]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef] [Green Version]
- CDC. Salmonella; CDC: Atlanta, GA, USA, 2020. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 10 June 2021).
- Snyder, T.R.; Boktor, S.W.; M’Ikanatha, N.M. Salmonellosis outbreaks by food vehicle, serotype, season, and geographical location, United States, 1998 to 2015. J. Food Prot. 2019, 82, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Boore, A.L.; Hoekstra, R.M.; Iwamoto, M.; Fields, P.I.; Bishop, R.D.; Swerdlow, D.L. Salmonella enterica infections in the United States and assessment of coefficients of variation: A novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS ONE 2015, 10, e0145416. [Google Scholar] [CrossRef] [Green Version]
- CDC. Outbreak of Salmonella Newport Infections Linked to Onions. Available online: https://www.cdc.gov/salmonella/newport-07-20/index.html (accessed on 28 May 2021).
- Kisluk, G.; Yaron, S. Presence and persistence of Salmonella enterica serotype Typhimurium in the phyllosphere and rhizosphere of spray-irrigated parsley. Appl. Environ. Microbiol. 2012, 78, 4030–4036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Biofilm formation and potential virulence factors of Salmonella strains isolated from ready-to-eat shrimps. PLoS ONE 2018, 13, e0204345. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.D.; Palmer, M.B.; Köster, W.L.; White, A.P. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Römling, U. Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef]
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella Biofilm Formation, Chronic Infection, and Immunity Within the Intestine and Hepatobiliary Tract. Front. Cell. Infect. Microbiol. 2020, 10, 624622. [Google Scholar] [CrossRef]
- Lories, B.; Belpaire, T.E.R.; Yssel, A.; Ramon, H.; Steenackers, H.P. Agaric acid reduces Salmonella biofilm formation by inhibiting flagellar motility. Biofilm 2020, 2, 100022. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Wang, Y.; Shivak, D.J.; Wong, C.S.; Hoffman, L.J.; Lam, S.; Kröger, C.; Cameron, A.D.; Townsend, H.G.; Köster, W.; et al. Bistable expression of CsgD in Salmonella enterica serovar Typhimurium connects virulence to persistence. Infect. Immun. 2015, 83, 2312–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamprokostopoulou, A.; Monteiro, C.; Rhen, M.; Römling, U. Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ. Microbiol. 2010, 12, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.; Pimentel-Filho, N.D.; Pinto, U.; Mantovani, H.; Oliveira, L.; Vanetti, M. Acyl homoserine lactone-based quorum sensing stimulates biofilm formation by Salmonella Enteritidis in anaerobic conditions. Arch. Microbiol. 2017, 199, 475–486. [Google Scholar] [CrossRef]
- Tursi, S.A.; Tükel, Ç. Curli-Containing Enteric Biofilms Inside and Out: Matrix Composition, Immune Recognition, and Disease Implications. Microbiol. Mol. Biol. Rev. 2018, 82, e00028-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dibb-Fuller, M.; Allen-Vercoe, E.; Thorns, C.; Woodward, M.J. Fimbriae-and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 1999, 145, 1023–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tursi, S.A.; Puligedda, R.D.; Szabo, P.; Nicastro, L.K.; Miller, A.L.; Qiu, C.; Gallucci, S.; Relkin, N.R.; Buttaro, B.A.; Dessain, S.K.; et al. Salmonella Typhimurium biofilm disruption by a human antibody that binds a pan-amyloid epitope on curli. Nat. Commun. 2020, 11, 1007. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.-W.; Sails, A. Molecular Medical Microbiology; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Favero, M.; Carson, L.; Bond, W.; Petersen, N. Pseudomonas aeruginosa: Growth in distilled water from hospitals. Science 1971, 173, 836–838. [Google Scholar] [CrossRef] [PubMed]
- CDC. Pseudomonas aeruginosa. Available online: https://www.cdc.gov/hai/organisms/pseudomonas.html (accessed on 28 May 2021).
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents—How P. aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [Green Version]
- Donelli, G. Biofilm-Based Healthcare-Associated Infections: Volume II; Springer: Berlin/Heidelberg, Germany, 2014; Volume 831, p. 3319097822. [Google Scholar] [CrossRef]
- Meliani, A.; Bensoltane, A. Review of Pseudomonas attachment and biofilm formation in food industry. Poult. Fish. Wildl. Sci. 2015, 3, 2–7. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, J.; Korber, D.; Hoyle, B.; Costerton, J.W.; Caldwell, D. Optical sectioning of microbial biofilms. J. Bacteriol. 1991, 173, 6558–6567. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Toutain, C.M.; Caizza, N.C.; Zegans, M.E.; O’Toole, G.A. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res. Microbiol. 2007, 158, 471–477. [Google Scholar] [CrossRef]
- Feldman, M.; Bryan, R.; Rajan, S.; Scheffler, L.; Brunnert, S.; Tang, H.; Prince, A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 1998, 66, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Craig, L.; Forest, K.T.; Maier, B. Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 2019, 17, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.; Koomey, M.; Maier, B. Dynamics of type IV pili is controlled by switching between multiple states. Biophys. J. 2009, 96, 1169–1177. [Google Scholar] [CrossRef] [Green Version]
- Ellison, C.K.; Kan, J.; Dillard, R.S.; Kysela, D.T.; Ducret, A.; Berne, C.; Hampton, C.M.; Ke, Z.; Wright, E.R.; Biais, N. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 2017, 358, 535–538. [Google Scholar] [CrossRef] [Green Version]
- Watts, T.H.; Scraba, D.G.; Paranchych, W. Formation of 9-nm filaments from pilin monomers obtained by octyl-glucoside dissociation of Pseudomonas aeruginosa pili. J. Bacteriol. 1982, 151, 1508–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, H.P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—A review. Gene 1997, 192, 99–108. [Google Scholar] [CrossRef]
- Chi, E.; Mehl, T.; Nunn, D.; Lory, S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect. Immun. 1991, 59, 822–828. [Google Scholar] [CrossRef] [Green Version]
- Comolli, J.C.; Waite, L.L.; Mostov, K.E.; Engel, J.N. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect. Immun. 1999, 67, 3207–3214. [Google Scholar] [CrossRef] [Green Version]
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008, 10, 2331–2343. [Google Scholar] [CrossRef]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- González, J.E.; Keshavan, N.D. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006, 70, 859–875. [Google Scholar] [CrossRef] [Green Version]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respirotoy Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [Green Version]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- Sakuragi, Y.; Kolter, R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5383–5386. [Google Scholar] [CrossRef] [Green Version]
- Vasseur, P.; Vallet-Gely, I.; Soscia, C.; Genin, S.; Filloux, A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 2005, 151, 985–997. [Google Scholar] [CrossRef] [Green Version]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, L.; Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004, 51, 675–690. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: From molecular biofilm biology to new treatment possibilities. APMIS 2014, 122, 1–51. [Google Scholar] [CrossRef]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef]
- Lydon, M.; Rahme, L.G. Production of Pseudomonas aeruginosa Intercellular small signaling molecules in human burn wounds. J. Pthog. 2011, 2011, 549302. [Google Scholar] [CrossRef]
- Xiao, G.; He, J.; Rahme, L.G. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 2006, 152, 1679–1686. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 2001, 98, 14613–14618. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, P. Putting an end to the Pseudomonas aeruginosa IQS controversy. Microologyopen 2020, 9, e962. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Yu, H.B.; Dela Ahator, S.; Wu, X.; Lv, S.; Zhang, L.H. Bacterial quorum-sensing signal IQS induces host cell apoptosis by targeting POT1–p53 signalling pathway. Cell. Microbiol. 2019, 21, e13076. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Goldsworthy, M.J.H. Gene expression of Pseudomonas aeruginosa and MRSA within a catheter-associated urinary tract infection biofilm model. BioSci. Horizon 2008, 1, 28–37. [Google Scholar] [CrossRef]



| Pathogen | Gram Stain | Spore Forming | Foods Involved | Infectious Dose | Disease Symptoms |
|---|---|---|---|---|---|
| Listeria monocytogenes | Positive | No | Ready-to-eat meat, dairy, fish, fruits, and vegetables. Foods with high protein content, such as deli meat, fish, and cheese. | <100–1011 CFU (colony forming unit) depending on individual immunological health [31,32]. | To healthy children and adults, flu-like symptoms include diarrhea, fever, vomiting, joint pain, headache. Invasive systemic disease in the immunocompromised host. Miscarriage and stillbirth in pregnant women. Meningitis or encephalitis in newborns and elderly. |
| Staphylococcus aureus | Positive | No | Milk products, meat, and hand-prepared foods. | S. aureus cells: 105–108 CFU/g. Toxin: 1 ng/g [33]. | Vomiting, diarrhea, and sometimes toxic shock symptoms including fever, low blood pressure, and even death. |
| Escherichia coli | Negative | No | Meat products, such as ground beef and sausage, fruits and vegetables. | Cause both food poisoning and infection. As low as 50–100 CFU of Enterohemorrhagic E. coli (EHEC) can cause infection [34]. | Vomiting, diarrhea, bloody diarrhea, hemorrhagic colitis, hemolytic uremic syndrome. |
| Salmonella enterica | Negative | No | Poultry products, meat, fish, vegetables, nuts, flours, milk, and drinking water. | Approximately 103–105 CFU is needed to cause diseases [35]. However, as low as 1–100 CFU is also implicated depending on the serovars involved [36]. | Typhoid fever, fever, vomiting, diarrhea, abdominal pain. It causes invasive disease in immunocompromised patients. |
| Pseudomonas aeruginosa | Negative | No | Not a common foodborne pathogen but may present in water, soil, plants, and foods. It contributes to the polymicrobial biofilm formation with other foodborne pathogens to be a food safety concern. | An opportunistic pathogen and infectious dose are highly variable; 103–109 CFU [37]. | Cause serious diseases in burn and cystic fibrosis patients with fever, chills, coughs with yellow, green, or bloody discharge. Gastroenteritis and diarrhea in some patients. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety. Foods 2021, 10, 2117. https://doi.org/10.3390/foods10092117
Bai X, Nakatsu CH, Bhunia AK. Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety. Foods. 2021; 10(9):2117. https://doi.org/10.3390/foods10092117
Chicago/Turabian StyleBai, Xingjian, Cindy H. Nakatsu, and Arun K. Bhunia. 2021. "Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety" Foods 10, no. 9: 2117. https://doi.org/10.3390/foods10092117