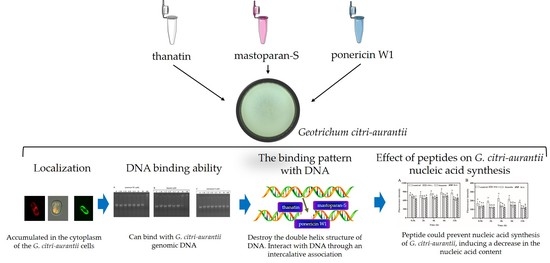
Interaction of Antimicrobial Peptide Ponericin W1, Thanatin, and Mastatopara-S with Geotrichum citri-aurantii Genomic DNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Pathogen Culture
2.3. Determining the Location of Peptides Using FITC Labelling
2.4. Preparation of G. citri-aurantii Genomic DNA
2.5. Gel Retardation Assay for Binding of Antimicrobial Peptides to G. citri-aurantii Genomic DNA
2.6. UV-Visible Spectra
2.7. Antimicrobial Peptides Competitively Bind DNA with Ethylene Diamine (EB)
2.8. Influence of Antimicrobial Peptides on DNA and RNA Synthesis in G. citri-aurantii
2.9. Statistical Analysis
3. Results
3.1. Localization of Peptides in G. citri-aurantii Conidia Cells
3.2. Gel Retardation Assay
3.3. UV-Visible Spectra
3.4. Competitive Binding of Peptides and EB with G. citri-aurantii DNA
3.5. Effect of Peptides on G. citri-aurantii Nucleic Acid Synthesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Shen, Y.; Chen, C.; Wan, C. Inhibition of Key Citrus Postharvest Fungal Strains by Plant Extracts In Vitro and In Vivo: A Review. Plants 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wang, W.; Deng, L.; Ming, J.; Yao, S.; Zeng, K. Control of sour rot in citrus fruit by three insect antimicrobial peptides. Postharvest Biol. Technol. 2019, 149, 200–208. [Google Scholar] [CrossRef]
- Kanetis, L.; Förster, H.; Adaskaveg, J.E. Comparative Efficacy of the New Postharvest Fungicides Azoxystrobin, Fludioxonil, and Pyrimethanil for Managing Citrus Green Mold. Plant Dis. 2007, 91, 1502–1511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, G.E. Efficacy of Guazatine and Iminoctadine for Control of Postharvest Decays of Oranges. Plant Dis. 1988, 72, 906. [Google Scholar] [CrossRef]
- Wang, Z. Indoor Toxicity Test of Giazatine on Geotrichum citri-aurantii in Main Citrus Producing Areas. Available online: https://d.wanfangdata.com.cn/thesis/D01879773 (accessed on 13 June 2019).
- Talibi, I.; Askarne, L.; Boubaker, H.; Boudyach, E.H.; Msanda, F.; Saadi, B.; Ait Ben Aoumar, A. Antifungal activity of some Moroccan plants against Geotrichum candidum, the causal agent of postharvest citrus sour rot. Crop Prot. 2012, 35, 41–46. [Google Scholar] [CrossRef]
- Palou, L.; Ali, A.; Fallik, E.; Romanazzi, G. GRAS, plant- and animal-derived compounds as alternatives to conventional fungicides for the control of postharvest diseases of fresh horticultural produce. Postharvest Biol. Technol. 2016, 122, 41–52. [Google Scholar] [CrossRef]
- Badosa, E.; Ferré, R.; Francés, J.; Bardají, E.; Feliu, L.; Planas, M.; Montesinos, E. Sporicidal activity of synthetic antifungal undecapeptides and control of Penicillium rot of apples. Appl. Environ. Microbiol. 2009, 75, 5563–5569. [Google Scholar] [CrossRef] [Green Version]
- Jia, F.; Wang, J.; Peng, J.; Zhao, P.; Kong, Z.; Wang, K.; Yan, W.; Wang, R. The in vitro, in vivo antifungal activity and the action mode of Jelleine-I against Candida species. Amino Acids 2018, 50, 229–239. [Google Scholar] [CrossRef]
- Waewthongrak, W.; Pisuchpen, S.; Leelasuphakul, W. Effect of Bacillus subtilis and chitosan applications on green mold (Penicilium digitatum Sacc. ) decay in citrus fruit. Postharvest Biol. Technol. 2015, 99, 44–49. [Google Scholar] [CrossRef]
- Yánez-Mendizábal, V.; Zeriouh, H.; Viñas, I.; Torres, R.; Usall, J.; Vicente, A.; de Pérez-García, A.; Teixidó, N. Biological control of peach brown rot (Monilinia spp.) by Bacillus subtilis CPA-8 is based on production of fengycin-like lipopeptides. Eur. J. Plant Pathol. 2012, 132, 609–619. [Google Scholar] [CrossRef]
- Shu, C.; Cui, K.; Li, Q.; Cao, J.; Jiang, W. Epsilon-poly-l-lysine (ε-PL) exhibits multifaceted antifungal mechanisms of action that control postharvest Alternaria rot. Int. J. Food Microbiol. 2021, 348, 109224. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Tang, Y.-L.; Shi, Y.-H.; Zhao, W.; Hao, G.; Le, G.-W. Interaction of MDpep9, a novel antimicrobial peptide from Chinese traditional edible larvae of housefly, with Escherichia coli genomic DNA. Food Chem. 2009, 115, 867–872. [Google Scholar] [CrossRef]
- Tomasinsig, L.; Scocchi, M.; Mettulio, R.; Zanetti, M. Genome-wide transcriptional profiling of the Escherichia coli response to a proline-rich antimicrobial peptide. Antimicrob. Agents Chemother. 2004, 48, 3260–3267. [Google Scholar] [CrossRef] [Green Version]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef] [Green Version]
- Orivel, J.; Redeker, V.; Le Caer, J.P.; Krier, F.; Revol-Junelles, A.M.; Longeon, A.; Chaffotte, A.; Dejean, A.; Rossier, J. Ponericins, new antibacterial and insecticidal peptides from the venom of the ant Pachycondyla goeldii. J. Biol. Chem. 2001, 276, 17823–17829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zare-Zardini, H.; Taheri-Kafrani, A.; Ordooei, M.; Ebrahimi, L.; Tolueinia, B.; Soleimanizadeh, M. Identification and biochemical characterization of a new antibacterial and antifungal peptide derived from the insect Sphodromantis viridis. Biochemistry 2015, 80, 433–440. [Google Scholar] [CrossRef]
- Omardien, S.; Drijfhout, J.W.; Zaat, S.A.; Brul, S. Cationic Amphipathic Antimicrobial Peptides Perturb the Inner Membrane of Germinated Spores Thus Inhibiting Their Outgrowth. Front. Microbiol. 2018, 9, 2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Louws, F.J.; Sutton, T.B.; Correll, J.C. A rapid qualitative molecular method for the identification of Colletotrichum acutatum and C. gloeosporioides. Eur. J. Plant Pathol. 2012, 132, 593–607. [Google Scholar] [CrossRef]
- Ning, Y.; Yan, A.; Yang, K.; Wang, Z.; Li, X.; Jia, Y. Antibacterial activity of phenyllactic acid against Listeria monocytogenes and Escherichia coli by dual mechanisms. Food Chem. 2017, 228, 533–540. [Google Scholar] [CrossRef]
- Imura, Y.; Nishida, M.; Ogawa, Y.; Takakura, Y.; Matsuzaki, K. Action mechanism of tachyplesin I and effects of PEGylation. Biochim. Biophys. Acta 2007, 1768, 1160–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, H.; Xie, M. Antibacterial mechanism of soybean isoflavone on Staphylococcus aureus. Arch. Microbiol. 2010, 192, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, Q.; Zhang, Q.; Huang, Y.; Su, Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Cao, Y. Fluorescence energy transfer between Acridine Orange and Safranine T and its application in the determination of DNA. Talanta 1999, 49, 377–383. [Google Scholar] [CrossRef]
- Sochacki, K.A.; Barns, K.J.; Bucki, R.; Weisshaar, J.C. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. USA 2011, 108, E77–E81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.-H.; Chen, C.; Jou, M.-L.; Lee, A.Y.-L.; Lin, Y.-C.; Yu, Y.-P.; Huang, W.-T.; Wu, S.-H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: Evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.-Y.; Xu, T.; Zheng, S.-T.; Zhang, L.-T.; Zhang, F.-C. Study on the interaction mechanism of antimicrobial peptide Cecropin-XJ in Xinjiang silkworm and Staphylococcus aureus DNA by spectra. Spectrosc. Spectr. Anal. 2008, 28, 612–616. [Google Scholar]
- Tian, T.; Hao, Y.; Sun, Y.; Xie, H.; Li, Z.; Liu, Q.L. Characteristics of A Novel Antimicrobial Peptide, Palustrin-2CE of Rana Chensinensis. In Proceedings of the The 12th Academic Conference on Biotoxin Research and Pharmaceutical Applications, Yan’an, China, 9–12 October 2015. [Google Scholar]
- Baranova, A.A.; Rogozhin, E.A.; Georgieva, M.L.; Bilanenko, E.N.; Kul’ko, A.B.; Yakushev, A.V.; Alferova, V.A.; Sadykova, V.S. Antimicrobial Peptides Produced by Alkaliphilic Fungi Emericellopsis alkalina: Biosynthesis and Biological Activity Against Pathogenic Multidrug-Resistant Fungi. Appl. Biochem. Microbiol. 2019, 55, 145–151. [Google Scholar] [CrossRef]
- Ferenc-András, B.; Zoltán-István, S.; Erika, S.; Pál, S.; Csongor, O.; Edit, S. Heterologous Expression and Purification of the Antimicrobial Peptide Buforin II. Bull. Med Sci. 2019, 92, 119–125. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Liu, S.; Li, X.; Wang, W.; Deng, L.; Zeng, K. Interaction of Antimicrobial Peptide Ponericin W1, Thanatin, and Mastatopara-S with Geotrichum citri-aurantii Genomic DNA. Foods 2021, 10, 1919. https://doi.org/10.3390/foods10081919
Zhang H, Liu S, Li X, Wang W, Deng L, Zeng K. Interaction of Antimicrobial Peptide Ponericin W1, Thanatin, and Mastatopara-S with Geotrichum citri-aurantii Genomic DNA. Foods. 2021; 10(8):1919. https://doi.org/10.3390/foods10081919
Chicago/Turabian StyleZhang, Hongyan, Sha Liu, Xindan Li, Wenjun Wang, Lili Deng, and Kaifang Zeng. 2021. "Interaction of Antimicrobial Peptide Ponericin W1, Thanatin, and Mastatopara-S with Geotrichum citri-aurantii Genomic DNA" Foods 10, no. 8: 1919. https://doi.org/10.3390/foods10081919