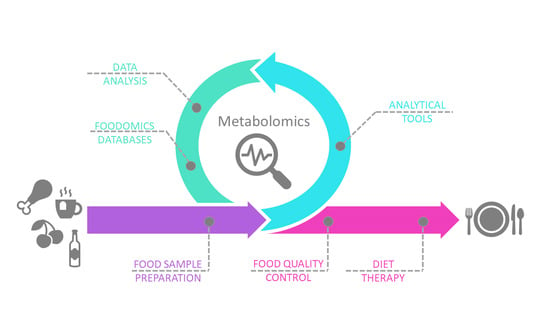
You Are What You Eat: Application of Metabolomics Approaches to Advance Nutrition Research
Abstract
:1. Introduction
2. (Un)Targeted Metabolomics
3. Metabolomics Analytical Tools
4. Foodomics Databases
5. Foodomics as a Standard for Safety and Quality Assessment
6. The Impact of Food Metabolites on Human Health
7. NMR in Analysis of Food Components
7.1. Tomato
7.2. Green Tea
7.3. Olive Oil
8. Metabolomics in Food Quality Control
9. Food Intake Promoting a Healthy Lifestyle
10. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hughes, M. Evolving Eating Habits as a Result of COVID-19. 2020. Available online: https://www.newfoodmagazine.com/article/109890/evolving-eating-habits-as-a-result-of-covid-19/ (accessed on 13 January 2021).
- Garneata, L.; Mircescu, G. Effect of Low-Protein Diet Supplemented With Keto Acids on Progression of Chronic Kidney Disease. J. Ren. Nutr. 2013, 23, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Bang, J.-K.; Kim, H.J.; Kim, J.-K.; Kim, Y.; Shin, S.Y. Antimicrobial specificity and mechanism of action of disulfide-removed linear analogs of the plant-derived Cys-rich antimicrobial peptide Ib-AMP1. Peptides 2009, 30, 2144–2149. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M.; Salek, R.M.; Griffin, J.L.; Merzaban, J.S. NMR-Based Metabolomics in Human Disease Diagnosis: Applications, Limitations, and Recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Emwas, A.M.; Luchinat, C.; Turano, P.; Tenori, L.; Roy, R.; Salek, R.M.; Ryan, D.; Merzaban, J.S.; Kaddurah-Daouk, R.; Zeri, A.C.; et al. Standardizing the Experimental Conditions for Using Urine in NMR-Based Metabolomic Studies with a Particular Focus on Diagnostic Studies: A Review. Metabolomics 2015, 11, 872–894. [Google Scholar] [CrossRef] [Green Version]
- Gowda, G.A.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-Based Methods for Early Disease Diagnostics. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-W.; Li, Q.-H.; Xu, Z.-D.; Dou, J.-J. Mass Spectrometry-Based Metabolomics in Health and Medical Science: A Systematic Review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef] [Green Version]
- Guasch-Ferré, M.; Bhupathiraju, S.N.; Hu, F.B. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin. Chem. 2018, 64, 82–98. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Zhu, Y.; Sang, S. A Novel LC-MS Based Targeted Metabolomic Approach to Study the Biomarkers of Food Intake. Mol. Nutr. Food Res. 2020, 64, e2000615. [Google Scholar] [CrossRef]
- Herrero, M.; Simó, C.; García-Cañas, V.; Ibáñez, E.; Cifuentes, A. Foodomics: MS-Based Strategies in Modern Food Science and Nutrition. Mass Spectrom. Rev. 2011, 31, 49–69. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Aliaga, I.; Kochhar, S.; Silva-Zolezzi, I. Biomarkers of Nutrient Bioactivity and Efficacy A Route toward Personalized Nutrition. J. Clin. Gastroenterol. 2012, 46, 545–554. [Google Scholar] [CrossRef]
- Kussmann, M.; Raymond, F.; Affolter, M. OMICS-Driven Biomarker Discovery in Nutrition and Health. J. Biotechnol. 2006, 124, 758–787. [Google Scholar] [CrossRef]
- Beckmann, M.; Lloyd, A.J.; Haldar, S.; Favé, G.; Seal, C.J.; Brandt, K.; Mathers, J.C.; Draper, J. Dietary Exposure Biomarker-Lead Discovery Based on Metabolomics Analysis of Urine Samples. Proc. Nutr. Soc. 2013, 72, 352–361. [Google Scholar] [CrossRef] [Green Version]
- Ibáñez, C.; Simó, C.; García-Cañas, V.; Cifuentes, A.; Castro-Puyana, M. Metabolomics, Peptidomics and Proteomics Applications of Capillary Electrophoresis-Mass Spectrometry in Foodomics: A Review. Anal. Chim. Acta 2013, 802, 1–13. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. Understanding ’Global’ Systems Biology: Metabonomics and the Continuum of Metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ’Metabonomics’: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Griffin, J.L. Metabolic Profiles to Define the Genome: Can We Hear the Phenotypes? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 857–871. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems Level Studies of Mammalian Metabolomes: The Roles of Mass Spectrometry and Nuclear Magnetic Resonance Spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef]
- Giordano, G.; Di Gangi, I.M.; Gucciardi, A.; Naturale, M. Quantification of Underivatised Amino Acids on Dry Blood Spot, Plasma, and Urine by HPLC–ESI–MS/MS. In Amino Acid Analysis: Methods and Protocols; Alterman, M.A., Hunziker, P., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 219–242. [Google Scholar]
- Lundström, S.L.; Saluja, R.; Adner, M.; Haeggström, J.Z.; Nilsson, G.; Wheelock, C.E. Lipid Mediator Metabolic Profiling Demonstrates Differences in Eicosanoid Patterns in Two Phenotypically Distinct Mast Cell Populations. J. Lipid Res. 2013, 54, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Gucciardi, A.; Pirillo, P.; Di Gangi, I.M.; Naturale, M.; Giordano, G. A Rapid UPLC–MS/MS Method for Simultaneous Separation of 48 Acylcarnitines in Dried Blood Spots and Plasma Useful as a Second-Tier Test for Expanded Newborn Screening. Anal. Bioanal. Chem. 2012, 404, 741–751. [Google Scholar] [CrossRef]
- Defernez, M.; Gunning, Y.M.; Parr, A.J.; Shepherd, L.V.T.; Davies, H.V.; Colquhoun, I.J. NMR and HPLC-UV Profiling of Potatoes with Genetic Modifications to Metabolic Pathways. J. Agric. Food Chem. 2004, 52, 6075–6085. [Google Scholar] [CrossRef]
- Al-Talla, Z.; Akrawi, S.H.; Emwas, A.-H.M. Solid State NMR and Bioequivalence Comparison of the Pharmacokinetic Parameters of Two Formulations of Clindamycin. Int. J. Clin. Pharmacol. Ther. 2011, 49, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Al-Talla, Z.A.; Akrawi, S.H.; Tolley, L.T.; Sioud, S.H.; Zaater, M.F.; Emwas, A.H.M. Bioequivalence Assessment of Two Formulations of Ibuprofen. Drug Des. Dev. Ther. 2011, 5, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Xie, H.-F.; Ma, Y.; Li, H.Y.; Li, C.P.; Chen, L.J.; Jiang, B.; Nian, B.; Guo, T.J.; Zhang, Z.Y.; et al. High Performance Liquid Chromatography and Metabolomics Analysis of Tannase Metabolism of Gallic Acid and Gallates in Tea Leaves. J. Agric. Food Chem. 2020, 68, 4946–4954. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, H.; Huang, X.; Guo, M.; Yang, Z.; Zhang, K.; Liu, J.; Wang, M.; Gong, Y.; Wei, J.; et al. Quality Assessment and Classification of Goji Berry by an HPLC-based Analytical Platform Coupled with Multivariate Statistical Analysis. Food Anal. Methods 2020, 13, 2222–2237. [Google Scholar] [CrossRef]
- Marchev, A.S.; Koycheva, I.K.; Aneva, I.Y.; Georgiev, M. Authenticity and Quality Evaluation of Different Rhodiola Species and Commercial Products Based on NMR-Spectroscopy and HPLC. Phytochem. Anal. 2020, 31, 756–769. [Google Scholar] [CrossRef]
- Ahmed, A.E.-S.I.; Wardell, J.N.; Thumser, A.E.; Avignone-Rossa, C.A.; Cavalli, G.; Hay, J.N.; Bushell, M.E. Metabolomic Profiling Can Differentiate between Bactericidal Effects of Free and Polymer Bound Halogen. J. Appl. Polym. Sci. 2011, 119, 709–718. [Google Scholar] [CrossRef] [Green Version]
- Scott, I.M.; Vermeer, C.P.; Liakata, M.; Corol, D.I.; Ward, J.L.; Lin, W.; Johnson, H.E.; Whitehead, L.; Kular, B.; Baker, J.M.; et al. Enhancement of Plant Metabolite Fingerprinting by Machine Learning. Plant Physiol. 2010, 153, 1506–1520. [Google Scholar] [CrossRef] [Green Version]
- Corte, L.; Rellini, P.; Roscini, L.; Fatichenti, F.; Cardinali, G. Development of a Novel, FTIR (Fourier Transform Infrared Spectroscopy) Based, Yeast Bioassay for Toxicity Testing and Stress Response Study. Anal. Chim. Acta 2010, 659, 258–265. [Google Scholar] [CrossRef]
- Gidman, E.; Goodacre, R.; Emmett, B.; Smith, A.R.; Gwynn-Jones, D. Investigating Plant-Plant Interference by Metabolic Fingerprinting. Phytochemistry 2003, 63, 705–710. [Google Scholar] [CrossRef]
- Lin, S.; Liu, N.; Yang, Z.; Song, W.; Wang, P.; Chen, H.; Lucio, M.; Schmitt-Kopplin, P.; Chen, G.; Cai, Z. GC/MS-Based Metabolomics Reveals Fatty Acid Biosynthesis and Cholesterol Metabolism in Cell Lines Infected with Influenza A Virus. Talanta 2010, 83, 262–268. [Google Scholar] [CrossRef]
- Riccio, M.F.; Saraiva, S.A.; Marques, L.A.; Alberici, R.; Haddad, R.; Möller, J.C.; Eberlin, M.N.; Catharino, R.R. Easy Mass Spectrometry for Metabolomics and Quality Control of Vegetable and Animal Fats. Eur. J. Lipid Sci. Technol. 2010, 112, 434–438. [Google Scholar] [CrossRef]
- Scheltema, R.; Decuypere, S.; Dujardin, J.; Watson, D.; Jansen, R.; Breitling, R. Simple Data-Reduction Method for High-Resolution LC–MS Data in Metabolomics. Bioanalysis 2009, 1, 1551–1557. [Google Scholar] [CrossRef] [Green Version]
- Wilson, I.D.; Plumb, R.; Granger, J.; Major, H.; Williams, R.; Lenz, E.M. HPLC-MS-based methods for the study of metabonomics. J. Chromatogr. B 2005, 817, 67–76. [Google Scholar] [CrossRef]
- Emwas, A.-H.M. The Strengths and Weaknesses of NMR Spectroscopy and Mass Spectrometry with Particular Focus on Metabolomics Research. In Metabonomics: Methods and Protocols; Bjerrum, J.T., Ed.; Springer: New York, NY, USA, 2015; pp. 161–193. [Google Scholar]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; AlAhmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Tiziani, S.; Emwas, A.-H.; Lodi, A.; Ludwig, C.; Bunce, C.M.; Viant, M.; Günther, U.L. Optimized Metabolite Extraction From Blood Serum For 1h Nuclear Magnetic Resonance Spectroscopy. Anal Biochem. 2008, 377, 16–23. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Ryan, D.; Brennan, L.; Tenori, L.; Luchinat, C.; Gao, X.; Zeri, A.C.; Gowda, G.A.N.; et al. Recommendations and Standardization of Biomarker Quantification Using NMR-Based Metabolomics with Particular Focus on Urinary Analysis. J. Proteome Res. 2016, 15, 360–373. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.R.; Kwon, H.N.; Nam, H.; Kim, J.J.; Park, S.; Kim, Y.-H. Urine-NMR Metabolomics for Screening of Advanced Colorectal Adenoma and Early Stage Colorectal Cancer. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Giraudeau, P. NMR-Based Metabolomics and Fluxomics: Developments and Future Prospects. Analyst 2020, 145, 2457–2472. [Google Scholar] [CrossRef]
- Chandra, K.; Harthi, S.; Almulhim, F.; Emwas, A.-H.; Jaremko, L.; Jaremko, M. The robust NMR Toolbox for Metabolomics. Mol. Omics 2021. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite Identification and Quantitation in LC-MS/MS-Based Metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhao, X.; Bai, C.; Zhao, C.; Lu, G.; Xu, G. LC–MS-Based Metabonomics Analysis. J. Chromatogr. B 2008, 866, 64–76. [Google Scholar] [CrossRef]
- Swartz, M.E. UPLC™: An Introduction and Review. J. Liq. Chromatogr. Relat. Technol. 2005, 28, 1253–1263. [Google Scholar] [CrossRef]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. Metabonomics Techniques and Applications to Pharmaceutical Research & Development. Pharm. Res. 2006, 23, 1075–1088. [Google Scholar]
- Grivet, J.-P.; Delort, A.-M. NMR for Microbiology: In vivo and In Situ Applications. Prog. Nucl. Magn. Spectrosc. 2009, 54, 1–53. [Google Scholar] [CrossRef]
- Grimes, J.H.; O’Connell, T.M. The Application of Micro-Coil NMR Probe Technology to Metabolomics of Urine and Serum. J. Biomol. NMR 2011, 49, 297–305. [Google Scholar] [CrossRef]
- Keun, H.C.; Beckonert, O.; Griffin, J.L.; Richter, C.; Moskau, D.; Lindon, J.C.; Nicholson, J.K. Cryogenic Probe 13C NMR Spectroscopy of Urine for Metabonomic Studies. Anal. Chem. 2002, 74, 4588–4593. [Google Scholar] [CrossRef]
- Ardenkjær-Larsen, J.H.; Fridlund, B.; Gram, A.; Hansson, G.; Lerche, M.H.; Servin, R.; Thaning, M.; Golman, K. Increase in Signal-to-Noise Ratio of >10,000 Times in Liquid-State NMR. Proc. Natl. Acad. Sci. USA 2003, 100, 10158–10163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, C.; Marin-Montesinos, I.; Saunders, M.G.; Emwas, A.-H.; Pikramenou, Z.; Hammond, S.P.; Günther, U.L. Application of Ex Situ Dynamic Nuclear Polarization in Studying Small Molecules. Phys. Chem. Chem. Phys. 2010, 12, 5868–5871. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Saunders, M.; Ludwig, C.; Günther, U.L. Determinants for Optimal Enhancement in Ex Situ DNP Experiments. Appl. Magn. Reson. 2008, 34, 483–494. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-Based Plant Metabolomics: Where Do We Stand, Where Do We Go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the Metabolome: Current Analytical Technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef]
- Holmes, E.; Tang, H.; Wang, Y.; Seger, C. The Assessment of Plant Metabolite Profiles by NMR-Based Methodologies. Planta Med. 2006, 72, 771–785. [Google Scholar] [CrossRef] [Green Version]
- Chandra, K.; Al-Harthi, S.; Sukumaran, S.; Almulhim, F.; Emwas, A.-H.; Atreya, H.S.; Jaremko, Ł.; Jaremko, M. NMR-Based Metabolomics with Enhanced Sensitivity. RSC Adv. 2021, 11, 8694–8700. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Alghrably, M.; Al-Harthi, S.; Poulson, B.G.; Szczepski, K.; Chandra, K.; Jaremko, M. New Advances in Fast Methods of 2D NMR Experiments. In Nuclear Magnetic Resonance; IntechOpen: London, UK, 2019; pp. 83–106. [Google Scholar]
- Salvino, R.A.; Colella, M.F.; De Luca, G. NMR-Based Metabolomics Analysis of Calabrian Citrus Fruit Juices and Its Application to Industrial Process Quality Control. Food Control. 2021, 121, 107619. [Google Scholar] [CrossRef]
- Cifani, C.; Alboni, S.; Mucci, A.; Benatti, C.; Botticelli, L.; Brunello, N.; Di Bonaventura, M.V.M.; Righi, V. serum metabolic signature of Binge-Like Palatable Food Consumption in Female Rats by Nuclear Magnetic Resonance Spectroscopy. NMR Biomed. 2021, 34, e4469. [Google Scholar] [CrossRef]
- Khodorova, N.V.; Rutledge, D.N.; Oberli, M.; Mathiron, D.; Marcelo, P.; Benamouzig, R.; Tome, D.; Gaudichon, C.; Pilard, S. Urinary Metabolomics Profiles Associated to Bovine Meat Ingestion in Humans. Mol. Nutr. Food Res. 2019, 63, e1700834. [Google Scholar] [CrossRef] [Green Version]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Lacueva, C.A.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, e1800384. [Google Scholar] [CrossRef]
- Kim, J.; Jung, Y.; Song, B.; Bong, Y.-S.; Ryu, D.H.; Lee, K.-S.; Hwang, G.-S. Discrimination of Cabbage (Brassica rapa spp. pekinensis) Cultivars Grown in Different Geographical Areas Using 1H NMR-Based Metabolomics. Food Chem. 2013, 137, 68–75. [Google Scholar] [CrossRef]
- Ritota, M.; Casciani, L.; Valentini, M. PGI Chicory (Cichorium intybus L.) Traceability by Means of HRMAS-NMR Spectroscopy: A Preliminary Study. J. Sci. Food Agric. 2012, 93, 1665–1672. [Google Scholar] [CrossRef]
- Takis, P.G.; Ghini, V.; Tenori, L.; Turano, P.; Luchinat, C. Uniqueness of the NMR Approach to Metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. [Google Scholar] [CrossRef]
- Marchand, J.; Martineau, E.; Guitton, Y.; Dervilly-Pinel, G.; Giraudeau, P. Multidimensional NMR Approaches Towards Highly Resolved, Sensitive and High-Throughput Quantitative Metabolomics. Curr. Opin. Biotechnol. 2017, 43, 49–55. [Google Scholar] [CrossRef]
- Féraud, B.; Govaerts, B.; Verleysen, M.; De Tullio, P. Statistical Treatment of 2D NMR COSY Spectra in Metabolomics: Data Preparation, Clustering-Based Evaluation of the Metabolomic Informative Content and Comparison with 1H-NMR. Metabolomics 2015, 11, 1756–1768. [Google Scholar] [CrossRef]
- Kruk, J.; Doskocz, M.; Jodłowska, E.; Zacharzewska, A.; Łakomiec, J.; Czaja, K.; Kujawski, J. NMR Techniques in Metabolomic Studies: A Quick Overview on Examples of Utilization. Appl. Magn. Reson. 2017, 48, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, C.; Viant, M.R. Two-Dimensional J-Resolved NMR Spectroscopy: Review of a Key Methodology in the Metabolomics Toolbox. Phytochem. Anal. 2009, 21, 22–32. [Google Scholar] [CrossRef]
- Bingol, K.; Brüschweiler, R. Multidimensional Approaches to NMR-Based Metabolomics. Anal. Chem. 2014, 86, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Donarski, J.; Jones, S.A.; Charlton, A.J. Application of Cryoprobe1H Nuclear Magnetic Resonance Spectroscopy and Multivariate Analysis for the Verification of Corsican Honey. J. Agric. Food Chem. 2008, 56, 5451–5456. [Google Scholar] [CrossRef]
- Sulaiman, F.; Azam, A.A.; Bustamam, M.S.A.; Fakurazi, S.; Abas, F.; Lee, Y.X.; Ismail, A.A.; Faudzi, S.M.M.; Ismail, I.S. Metabolite Profiles of Red and Yellow Watermelon (Citrullus lanatus) Cultivars Using a 1H-NMR Metabolomics Approach. Molecules 2020, 25, 3235. [Google Scholar] [CrossRef]
- Cho, I.H.; Kim, Y.-S.; Choi, H.-K. Metabolomic Discrimination of Different Grades of Pine-Mushroom (Tricholoma matsutake Sing.) Using 1h NMR Spectrometry and Multivariate Data Analysis. J. Pharm. Biomed. Anal. 2007, 43, 900–904. [Google Scholar] [CrossRef]
- Son, H.-S.; Kim, K.M.; Berg, F.V.D.; Hwang, G.-S.; Park, W.-M.; Lee, C.-H.; Hong, Y.-S. 1H Nuclear Magnetic Resonance-Based Metabolomic Characterization of Wines by Grape Varieties and Production Areas. J. Agric. Food Chem. 2008, 56, 8007–8016. [Google Scholar] [CrossRef]
- Son, H.-S.; Hwang, G.-S.; Ahn, H.-J.; Park, W.-M.; Lee, C.-H.; Hong, Y.-S. Characterization of Wines from Grape Varieties through Multivariate Statistical Analysis of 1h NMR Spectroscopic Data. Food Res. Int. 2009, 42, 1483–1491. [Google Scholar] [CrossRef]
- Pongsuwan, W.; Bamba, T.; Harada, K.; Yonetani, T.; Kobayashi, A.; Fukusaki, E. High-Throughput Technique for Comprehensive Analysis of Japanese Green Tea Quality Assessment Using Ultra-performance Liquid Chromatography with Time-of-Flight Mass Spectrometry (UPLC/TOF MS). J. Agric. Food Chem. 2008, 56, 10705–10708. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Kim, H.K.; Lefeber, A.W.; Erkelens, C.; Angelova, N.; Choi, Y.H.; Verpoorte, R. Application of Two-Dimensional Nuclear Magnetic Resonance Spectroscopy to Quality Control of Ginseng Commercial Products. Planta Med. 2006, 72, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Bearden, D.W.; Bundy, J.G.; Burton, I.W.; Collette, T.W.; Ekman, D.R.; Ezernieks, V.; Karakach, T.K.; Lin, C.Y.; Rochfort, S.; et al. International NMR-Based Environmental Metabolomics Intercomparison Exercise. Environ. Sci. Technol. 2009, 43, 219–225. [Google Scholar] [CrossRef]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M. High-Throughput Tissue Extraction Protocol for NMR- and MS-Based Metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef]
- Barros, E.; Lezar, S.; Anttonen, M.J.; Van Dijk, J.P.; Röhlig, R.M.; Kok, E.J.; Engel, K.-H. Comparison of two GM Maize Varieties with a Near-Isogenic Non-GM Variety Using Transcriptomics, Proteomics and Metabolomics. Plant Biotechnol. J. 2010, 8, 436–451. [Google Scholar] [CrossRef]
- Kim, H.-S.; Park, S.J.; Hyun, S.-H.; Yang, S.-O.; Lee, J.; Auh, J.-H.; Kim, J.-H.; Cho, S.-M.; Marriott, P.J.; Choi, H.-K. Biochemical Monitoring of Black Raspberry (Rubus coreanus Miquel) Fruits According to Maturation Stage by 1 H NMR Using Multiple Solvent Systems. Food Res. Int. 2011, 44, 1977–1987. [Google Scholar] [CrossRef]
- Le Gall, G.; Dupont, M.S.; Mellon, F.A.; Davis, A.L.; Collins, G.J.; Verhoeyen, M.E.; Colquhoun, I.J. Characterization and Content of Flavonoid Glycosides in Genetically Modified Tomato (Lycopersicon esculentum) Fruits. J. Agric. Food Chem. 2003, 51, 2438–2446. [Google Scholar] [CrossRef]
- Le Gall, G.; Colquhoun, I.J.; Davis, A.L.; Collins, G.J.; Verhoeyen, M.E. Metabolite Profiling of Tomato (Lycopersicon esculentum) Using 1H NMR Spectroscopy as a Tool to Detect Potential Unintended Effects Following a Genetic Modification. J. Agric. Food Chem. 2003, 51, 2447–2456. [Google Scholar] [CrossRef]
- Tiziani, S.; Schwartz, S.J.; Vodovotz, Y. Profiling of Carotenoids in Tomato Juice by One- and Two-Dimensional NMR. J. Agric. Food Chem. 2006, 54, 6094–6100. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Sobolev, A.P.; Neelam, A.; Goyal, R.K.; Handa, A.K.; Segre, A.L. Nuclear Magnetic Resonance Spectroscopy-Based Metabolite Profiling of Transgenic Tomato Fruit Engineered to Accumulate Spermidine and Spermine Reveals Enhanced Anabolic and Nitrogen-Carbon Interactions. Plant Physiol. 2006, 142, 1759–1770. [Google Scholar] [CrossRef] [Green Version]
- Sobolev, A.P.; Segre, A.; Lamanna, R. Proton High-Field NMR Study of Tomato Juice. Magn. Reson. Chem. 2003, 41, 237–245. [Google Scholar] [CrossRef]
- Moco, S.; Forshed, J.; De Vos, R.C.H.; Bino, R.J.; Vervoort, J. Intra- and Inter-Metabolite Correlation Spectroscopy of Tomato Metabolomics Data Obtained by Liquid Chromatography-Mass Spectrometry and Nuclear Magnetic Resonance. Metabolomics 2008, 4, 202–215. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, B.; Cloarec, O.; Tang, H.; Staerk, D.; Jaroszewski, J. Multivariate Analysis of Integrated and Full-Resolution 1 H-NMR Spectral Data from Complex Pharmaceutical Preparations: St. John’s Wort. Planta Med. 2006, 72, 556–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, G.; Röseler, C.; Büter, K.B.; Simmen, U. Classification and Correlation of St. John’s Wort Extracts by Nuclear Magnetic Resonance Spectroscopy, Multivariate Data Analysis and Pharmacological Activity. Planta Med. 2004, 70, 771–777. [Google Scholar] [CrossRef]
- Agnolet, S.; Jaroszewski, J.W.; Verpoorte, R.; Staerk, D. 1H NMR-Based Metabolomics Combined with HPLC-PDA-MS-SPE-NMR for Investigation of Standardized Ginkgo biloba Preparations. Metabolomics 2010, 6, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes, A. Advanced Separation Methods in Food Analysis. J. Chromatogr. A 2009, 1216, 7109–7358. [Google Scholar] [CrossRef] [Green Version]
- Kudo, T.; Kobayashi, M.; Terashima, S.; Katayama, M.; Ozaki, S.; Kanno, M.; Saito, M.; Yokoyama, K.; Ohyanagi, H.; Aoki, K.; et al. TOMATOMICS: A Web Database for Integrated Omics Information in Tomato. Plant Cell Physiol. 2017, 58, e8. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A Dynamic Web Platform for Genome Visualization and Analysis. Genome Biol. 2016, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Shikata, M.; Hoshikawa, K.; Ariizumi, T.; Fukuda, N.; Yamazaki, Y.; Ezura, H. TOMATOMA Update: Phenotypic and Metabolite Information in the Micro-Tom Mutant Resource. Plant Cell Physiol. 2015, 57, e11. [Google Scholar] [CrossRef] [Green Version]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A Novel Tomato Mutant Database Distributing Micro-Tom Mutant Collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Chu, G.-X.; Liu, X.-S.; Tang, X.; Wang, W.; Liu, G.-J.; Yang, T.; Ling, T.-J.; Wang, X.-G.; Zhang, Z.-Z.; et al. TMDB: A Literature-Curated Database for Small Molecular Compounds Found from Tea. BMC Plant Biol. 2014, 14, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Foroutan, A.; Fitzsimmons, C.; Mandal, R.; Piri-Moghadam, H.; Zheng, J.; Guo, A.; Li, C.; Guan, L.L.; Wishart, D.S. The Bovine Metabolome. Metabolites 2020, 10, 233. [Google Scholar] [CrossRef]
- Foroutan, A.; Guo, A.C.; Vazquez-Fresno, R.; Lipfert, M.; Zhang, L.; Zheng, J.; Badran, H.; Budinski, Z.; Mandal, R.; Ametaj, B.N.; et al. Chemical Composition of Commercial Cow’s Milk. J. Agric. Food Chem. 2019, 67, 4897–4914. [Google Scholar] [CrossRef]
- Bozkurt, F.; Tekin, R.; Gulsun, S.; Satıcı, Ö.; Deveci, O.; Hosoglu, S. The Levels of Copper, Zinc and Magnesium in Type II Diabetic Patients Complicated with Foot Infections. Int. J. Diabetes Dev. Ctries. 2013, 33, 165–169. [Google Scholar] [CrossRef]
- Forte, G.; Bocca, B.; Senofonte, O.; Petrucci, F.; Brusa, L.; Stanzione, P.; Zannino, S.; Violante, N.; Alimonti, A.; Sancesario, G. Trace and Major Elements in Whole Blood, Serum, Cerebrospinal Fluid and Urine of Patients with Parkinson’s Disease. J. Neural Transm. 2004, 111, 1031–1040. [Google Scholar] [CrossRef]
- Namkung, Y.; Jang, S. Does Food Quality Really Matter in Restaurants? Its Impact on Customer Satisfaction and Behavioral Intentions. J. Hosp. Tour. Res. 2007, 31, 387–409. [Google Scholar] [CrossRef]
- Baiardi, D.; Puglisi, R.; Scabrosetti, S. Individual Attitudes on Food Quality and Safety: Empirical Evidence on EU Countries. Food Qual. Prefer. 2016, 49, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Grujić, S.; Grujić, R.; Petrović, Đ.; Gajić, J. Knowledge of Food Quality and Additives and Its Impact on Food Preference. Acta Sci. Pol. Technol. Aliment. 2013, 12, 215–222. [Google Scholar]
- Mergenthaler, M.; Weinberger, K.; Qaim, M. Consumer Valuation of Food Quality and Food Safety Attributes in Vietnam. Rev. Agric. Econ. 2009, 31, 266–283. [Google Scholar] [CrossRef]
- Manig, C.; Moneta, A. More or Better? Measuring Quality Versus Quantity in Food Consumption. J. Bioeconomics 2013, 16, 155–178. [Google Scholar] [CrossRef] [Green Version]
- Roser, M.; Ortiz-Ospina, E. Global Rise of Education. Available online: https://www.semanticscholar.org/paper/Global-Rise-of-Education-Roser-Ortiz-Ospina/57319c871ba3e3c858f159182ba91e83c0c202c6?p2df (accessed on 13 January 2021).
- Ross, C.E.; Wu, C.-L. The Links Between Education and Health. Am. Sociol. Rev. 1995, 60, 719–745. [Google Scholar] [CrossRef]
- Cutler, D.; Lleras-Muney, A. Education and Health: Evaluating Theories and Evidence. Available online: https://www.nber.org/system/files/working_papers/w12352/w12352.pdf (accessed on 13 January 2021).
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. MEATabolomics: Muscle and Meat Metabolomics in Domestic Animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef]
- Glitsch, K. Consumer Perceptions of Fresh Meat Quality: Cross-National Comparison. Br. Food J. 2000, 102, 177–194. [Google Scholar] [CrossRef]
- Wideman, N.; O’Bryan, C.; Crandall, P. Factors Affecting Poultry Meat Colour and Consumer Preferences—A Review. World’s Poult. Sci. J. 2016, 72, 353–366. [Google Scholar] [CrossRef]
- Seideman, S.C.; Cross, H.R.; Smith, G.C.; Durland, P.R. Factors Associated with Fresh Meat Color: A Review. J. Food Qual. 1984, 6, 211–237. [Google Scholar] [CrossRef]
- Suman, S.P.; Joseph, P. Myoglobin Chemistry and Meat Color. Annu. Rev. Food Sci. Technol. 2013, 4, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, R.; Kiyimba, F.; Gonzalez, J.M.; Mafi, G.G.; DeSilva, U. Impact of Up- and Downregulation of Metabolites and Mitochondrial Content on pH and Color of the Longissimus Muscle from Normal-pH and Dark-Cutting Beef. J. Agric. Food Chem. 2020, 68, 7194–7203. [Google Scholar] [CrossRef]
- Beauclercq, S.; Nadal-Desbarats, L.; Hennequet-Antier, C.; Collin, A.; Tesseraud, S.; Bourin, M.; Le Bihan-Duval, E.; Berri, C. Serum and Muscle Metabolomics for the Prediction of Ultimate pH, a Key Factor for Chicken-Meat Quality. J. Proteome Res. 2016, 15, 1168–1178. [Google Scholar] [CrossRef]
- Guo, Y.; Bian, X.; Liu, J.; Zhu, M.; Li, L.; Yao, T.; Tang, C.; Ravichandran, V.; Liao, P.; Papadimitriou, K.; et al. Dietary Components, Microbial Metabolites and Human Health: Reading between the Lines. Foods 2020, 9, 1045. [Google Scholar] [CrossRef]
- Roager, H.M.; Dragsted, L.O. Diet-Derived Microbial Metabolites in Health and Disease. Nutr. Bull. 2019, 44, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [Green Version]
- Butel, M.-J. Probiotics, Gut Microbiota and Health. Méd. Mal. Infect. 2014, 44, 1–8. [Google Scholar] [CrossRef]
- Danneskiold-Samsøe, N.B.; Barros, H.D.D.F.Q.; Santos, R.; Bicas, J.L.; Cazarin, C.B.B.; Madsen, L.; Kristiansen, K.; Pastore, G.M.; Brix, S.; Júnior, M.R.M. Interplay between Food and Gut Microbiota in Health and Disease. Food Res. Int. 2019, 115, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Farag, M.A.; Abdelwareth, A.; Sallam, I.E.; Shorbagi, M.; Jehmlich, N.; Fritz-Wallace, K.; Schäpe, S.S.; Rolle-Kampczyk, U.; Ehrlich, A.; Wessjohann, L.A.; et al. Metabolomics Reveals Impact of Seven Functional Foods on Metabolic Pathways in a Gut Microbiota Model. J. Adv. Res. 2020, 23, 47–59. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Gu, Q.; Li, P. Biosynthesis of Vitamins by Probiotic Bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; Rao, V., Rao, L., Eds.; IntechOpen: London, UK, 2016. [Google Scholar]
- Hasan, N.; Yang, H. Factors Affecting the Composition of the Gut Microbiota, and Its Modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef] [Green Version]
- Andersen, M.-B.S.; Rinnan, Å.; Manach, C.; Poulsen, S.K.; Pujos-Guillot, E.; Larsen, T.M.; Astrup, A.; Dragsted, L.O. Untargeted Metabolomics as a Screening Tool for Estimating Compliance to a Dietary Pattern. J. Proteome Res. 2014, 13, 1405–1418. [Google Scholar] [CrossRef]
- Posma, J.M.; Garcia-Perez, I.; Frost, G.; Aljuraiban, G.S.; Chan, Q.; Van Horn, L.; Daviglus, M.; Stamler, J.; Holmes, E.; Elliott, P.; et al. Nutriome–Metabolome Relationships Provide Insights into Dietary Intake and Metabolism. Nat. Food 2020, 1, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, J.; Ishikawa, T.; Satoh, M. Polar Constituents of Celery seed. Phytochemistry 2003, 64, 1003–1011. [Google Scholar] [CrossRef]
- Pérez, E.M.S.; Iglesias, M.J.; López-Ortiz, F.; Pérez, I.S.; Galera, M.M. Study of the Suitability of Hrmas NMR for Metabolic Profiling of Tomatoes: Application to Tissue Differentiation and Fruit Ripening. Food Chem. 2010, 122, 877–887. [Google Scholar] [CrossRef]
- Pérez, E.M.S.; López, J.G.; Iglesias, M.J.; Ortiz, F.L.; Toresano, F.; Camacho, F. HRMAS-Nuclear Magnetic Resonance Spectroscopy Characterization of Tomato “Flavor Varieties” from Almería (Spain). Food Res. Int. 2011, 44, 3212–3221. [Google Scholar] [CrossRef]
- Stark, R.E.; Yan, B.; Stanley-Fernandez, S.M.; Chen, Z.-J.; Garbow, J.R. NMR Characterization of Hydration and Thermal Stress in Tomato Fruit Cuticles. Phytochemistry 2008, 69, 2689–2695. [Google Scholar] [CrossRef]
- de Falco, B.; Manzo, D.; Incerti, G.; Garonna, A.P.; Ercolano, M.; Lanzotti, V. Metabolomics Approach Based on NMR Spectroscopy and Multivariate Data Analysis to Explore the Interaction between the Leafminer Tuta absoluta and Tomato (Solanum lycopersicum). Phytochem. Anal. 2019, 30, 556–563. [Google Scholar] [CrossRef]
- Meza, S.L.R.; Egea, I.; Massaretto, I.L.; Morales, B.; Purgatto, E.; Egea-Fernández, J.M.; Bolarin, M.C.; Flores, F.B. Traditional Tomato Varieties Improve Fruit Quality Without Affecting Fruit Yield Under Moderate Salt Stress. Front. Plant Sci. 2020, 11, 11. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Chen, P.; Harnly, J.M. New Phenolic Components and Chromatographic Profiles of Green and Fermented Teas. J. Agric. Food Chem. 2008, 56, 8130–8140. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-E.; Lee, B.-J.; Chung, J.-O.; Kim, H.-N.; Kim, E.-H.; Jung, S.; Lee, H.; Lee, S.-J.; Hong, Y.-S. Metabolomic Unveiling of a Diverse Range of Green Tea (Camellia sinensis) Metabolites Dependent on Geography. Food Chem. 2015, 174, 452–459. [Google Scholar] [CrossRef]
- Lee, J.-E.; Lee, B.-J.; Hwang, J.-A.; Ko, K.-S.; Chung, J.-O.; Kim, E.-H.; Lee, S.-J.; Hong, Y.-S. Metabolic Dependence of Green Tea on Plucking Positions Revisited: A Metabolomic Study. J. Agric. Food Chem. 2011, 59, 10579–10585. [Google Scholar] [CrossRef]
- Napolitano, J.; Gödecke, T.; Lankin, D.C.; Jaki, B.U.; McAlpine, J.B.; Chen, S.-N.; Pauli, G.F. Orthogonal Analytical Methods for Botanical Standardization: Determination of Green Tea Catechins by qNMR and LC–MS/MS. J. Pharm. Biomed. Anal. 2014, 93, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-E.; Lee, B.-J.; Chung, J.-O.; Shin, H.-J.; Lee, S.-J.; Lee, C.-H.; Hong, Y.-S. 1H NMR-Based Metabolomic Characterization During Green Tea (Camellia sinensis) Fermentation. Food Res. Int. 2011, 44, 597–604. [Google Scholar] [CrossRef]
- Wahyuni, D.S.C.; Kristanti, M.W.; Putri, R.K.; Rinanto, Y. NMR Metabolic Profiling of Green Tea (Camellia sinensis L.) Leaves Grown at Kemuning, Indonesia. J. Phys. Conf. Ser. 2017, 795, 12013. [Google Scholar] [CrossRef]
- Morelló, J.-R.; Romero, M.-P.; Ramo, T.; Motilva, M.-J. Evaluation of L-Phenylalanine Ammonia-Lyase Activity and Phenolic Profile in Olive Drupe (Olea europaea L.) from Fruit Setting Period to Harvesting Time. Plant Sci. 2005, 168, 65–72. [Google Scholar] [CrossRef]
- Nguyen, X.H.T.; Juvik, O.J.; Øvstedal, D.O.; Fossen, T. 6-Carboxydihydroresveratrol 3-O-β-Glucopyranoside—A Novel Natural Product from the Cretaceous Relict Metasequoia glyptostroboides. Fitoterapia 2014, 95, 109–114. [Google Scholar] [CrossRef]
- Hashmi, M.A.; Khan, A.; Hanif, M.; Farooq, U.; Perveen, S. Traditional Uses, Phytochemistry, and Pharmacology of Olea europaea (Olive). Evid. Based Complement. Altern. Med. 2015, 2015, 541591. [Google Scholar] [CrossRef] [Green Version]
- Erbay, Z.; Icier, F. The Importance and Potential Uses of Olive Leaves. Food Rev. Int. 2010, 26, 319–334. [Google Scholar] [CrossRef]
- Beteinakis, S.; Papachristodoulou, A.; Gogou, G.; Katsikis, S.; Mikros, E.; Halabalaki, M. NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters. Molecules 2020, 25, 3339. [Google Scholar] [CrossRef]
- Merchak, N.; El Bacha, E.; Khouzam, R.B.; Rizk, T.; Akoka, S.; Bejjani, J. Geoclimatic, Morphological, and Temporal Effects on Lebanese Olive Oils Composition and Classification: A 1H NMR Metabolomic Study. Food Chem. 2017, 217, 379–388. [Google Scholar] [CrossRef]
- Rongai, D.; Sabatini, N.; Del Coco, L.; Perri, E.; Del Re, P.; Simone, N.; Marchegiani, D.; Fanizzi, F.P. 1H NMR and Multivariate Analysis for Geographic Characterization of Commercial Extra Virgin Olive Oil: A Possible Correlation with Climate Data. Foods 2017, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- Rongai, D.; Del Re, P.; Simone, N.; Sabatini, N. 13C NMR Analysis for the Geographical Characterization of Extra Virgin Olive Oil (EVOO) Produced in Some Italian Regions. Nutr. Food Sci. Int. J. 2020, 8, 79–82. [Google Scholar]
- Consonni, R.; Cagliani, L.R. Geographical Characterization of Polyfloral and Acacia Honeys by Nuclear Magnetic Resonance and Chemometrics. J. Agric. Food Chem. 2008, 56, 6873–6880. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Salces, R.M.; Moreno-Rojas, J.M.; Holland, M.V.; Reniero, F.; Guillou, C.; Héberger, K. Virgin Olive Oil Authentication by Multivariate Analyses of1H NMR Fingerprints and δ13C and δ2H Data. J. Agric. Food Chem. 2010, 58, 5586–5596. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.; Capitani, D. NMR and Chemometrics in Tracing European Olive Oils: The Case Study of Ligurian Samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Donarski, J.; Jones, S.A.; Harrison, M.; Driffield, M.; Charlton, A.J. Identification of Botanical Biomarkers Found in Corsican Honey. Food Chem. 2010, 118, 987–994. [Google Scholar] [CrossRef]
- Santos, P.; Filho, E.P.; Rodriguez-Saona, L. Rapid Detection and Quantification of Milk Adulteration Using Infrared Microspectroscopy and Chemometrics Analysis. Food Chem. 2013, 138, 19–24. [Google Scholar] [CrossRef]
- Osorio, M.; Moloney, A.; Brennan, L.; Monahan, F. Authentication of Beef Production Systems Using a Metabolomic-Based Approach. Animal 2012, 6, 167–172. [Google Scholar] [CrossRef]
- Kanoski, S.E.; Davidson, T.L. Western Diet Consumption and Cognitive Impairment: Links to Hippocampal Dysfunction and Obesity. Physiol. Behav. 2011, 103, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Davidson, T.; Hargrave, S.; Swithers, S.; Sample, C.; Fu, X.; Kinzig, K.; Zheng, W. Inter-Relationships Among Diet, Obesity and Hippocampal-Dependent Cognitive Function. Neuroscience 2013, 253, 110–122. [Google Scholar] [CrossRef]
- Tuck, C.J.; Staudacher, H.M. The Keto Diet and the Gut: Cause for Concern? Lancet Gastroenterol. Hepatol. 2019, 4, 908–909. [Google Scholar] [CrossRef]
- Gogou, M.; Kolios, G. Are Therapeutic Diets an Emerging Additional Choice in Autism Spectrum Disorder Management? World J. Pediatr. 2018, 14, 215–223. [Google Scholar] [CrossRef]
- Olson, C.A.; Vuong, H.E.; Yano, J.M.; Liang, Q.Y.; Nusbaum, D.; Hsiao, E.Y. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018, 173, 1728–1741.e13. [Google Scholar] [CrossRef] [Green Version]
- Zupec-Kania, B.; Zupanc, M.L. Long-Term Management of the Ketogenic Diet: Seizure Monitoring, Nutrition, and Supplementation. Epilepsia 2008, 49, 23–26. [Google Scholar] [CrossRef]
- Gutiérrez-Repiso, C.; Hernández-García, C.; García-Almeida, J.M.; Bellido, D.; Martín-Núñez, G.M.; Sánchez-Alcoholado, L.; Alcaide-Torres, J.; Sajoux, I.; Tinahones, F.J.; Moreno-Indias, I. Effect of Synbiotic Supplementation in a Very-Low-Calorie Ketogenic Diet on Weight Loss Achievement and Gut Microbiota: A Randomized Controlled Pilot Study. Mol. Nutr. Food Res. 2019, 63, e1900167. [Google Scholar] [CrossRef]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic Diet and Microbiota: Friends or Enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef] [Green Version]
- Berkow, S.E.; Barnard, N.; Eckart, J.; Katcher, H. Four Therapeutic Diets: Adherence and Acceptability. Can. J. Diet. Pr. Res. 2010, 71, 199–204. [Google Scholar] [CrossRef]
- Long, T.; Wagner, L. Therapeutic Diets. In Integrative and Functional Medical Nutrition Therapy: Principles and Practices; Noland, D., Drisko, J.A., Wagner, L., Eds.; Springer: Cham, Switzerland, 2020; pp. 743–754. [Google Scholar]
- Licha, D.; Vidali, S.; Aminzadeh-Gohari, S.; Alka, O.; Breitkreuz, L.; Kohlbacher, O.; Reischl, R.J.; Feichtinger, R.G.; Kofler, B.; Huber, C.G. Untargeted Metabolomics Reveals Molecular Effects of Ketogenic Diet on Healthy and Tumor Xenograft Mouse Models. Int. J. Mol. Sci. 2019, 20, 3873. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Black, A.; Kales, S.N.; Vattem, D.; Ruiz-Canela, M.; Sotos-Prieto, M. Metabolomics and Microbiomes as Potential Tools to Evaluate the Effects of the Mediterranean Diet. Nutrients 2019, 11, 207. [Google Scholar] [CrossRef] [Green Version]


| Analytical Method | Example of Use in Sample Analysis | Purpose of Study | Reference |
|---|---|---|---|
| Honey | Determination of the geographical origin of honey | [70] | |
| Watermelon | Description of metabolites in red and yellow watermelon cultivars with focus on the carotenoid group | [71] | |
| Pine mushrooms | Ilustration of differences among the various grades of pine mushroom | [72] | |
| Wine | Characterization of the metabolites in wines vinified in different continental areas and from different grape varieties to improve the quality of wine | [73,74] | |
| Green tea | Developing a method to evaluate the quality of Japanese green tea | [75] | |
| Ginseng (Panax ginseng) | Developing a method to control the quality of Ginseng commercial products | [76] | |
| NMR | Fish tissue | Demonstrating the efficacy of NMR in environmental research/Optimization of tissue extraction methods | [77,78] |
| Maize (Zea mays) | Safety evaluation of genetically modified maize | [79] | |
| Black raspberry fruit | Monitoring biochemical changes of black raspberry fruits based on the maturation level, extraction method and NMR-solvent conditions | [80] | |
| Tomato dry-powder with organic solvents | Profile and level characterization of carotenoids and flavonoid glycosides/Detection of potential unintended effects of genetic modification on tomato | [81,82,83] | |
| Tomato dry-powder with water | Evaluation of the role of polyamines in growth and development in tomato | [84] | |
| Tomato juice/pulp | Metabolite characterization of tomato juice and pulp/Metabolite profiling of 50 different tomato cultivars | [85,86] | |
| St. John’s wort | Comparison of St. John’s wort extracts that have been subjected to the same standardization procedure/Prediction of pharmacological efficacy of different extracts | [87,88] | |
| HPLC-NMR | Gingko (Gingko biloba) | Investigation of metabolomic composition of 16 commercially available Ginkgo preparations and identification of flavonoid glycosides and terpene trilactones | [89] |
| Food | Study | Analytical Method | Statistical Analysis | References |
|---|---|---|---|---|
| Olive oil | 1H NMR fingerprints of virgin olive oils (VOOs) from the Mediterranean basin (three harvests) were analyzed by principal component analysis (PCA), linear discriminant analysis (LDA), and partial least-squares discriminant analysis (PLS-DA) to determine their geographical origin at the national, regional, or PDO level | 1H- and 13C-NMR | ANOVA PCA LDA PLS-DA | [150] |
| Olive oil | NMR-based metabolomics approach used to classify the extra virgin olive oils based on their geographical origin | 1H-NMR | PLS-DA SIMCA | [151] |
| Honey | NMR was used as an analytical tool to confirm honey origin | 1H-NMR | -- | [152] |
| Honey | Proton NMR was used in conjunction with multivariate analysis techniques (PLS, PLS-LDA) to classify honey into groups by geographical origin including honey from Corsica Island and other part of Europe | 1H-NMR | PLS PLS-LDA PLS-GP | [70] |
| Honey | 41 honey samples from different countries were analyzed using 1H-NMR spectroscopy in conjunction with PCA to distinguish between polyfloral and acacia honey samples | 1H-NMR | PCA PLS-DA | [149] |
| Milk | The application of attenuated total reflectance mid-infrared (MIR) microspectroscopy was evaluated as a rapid method for detection and quantification of milk adulteration. Milk samples were purchased from local grocery stores (Columbus, OH, USA) and spiked at different concentrations with whey, hydrogen peroxide, synthetic urine, urea and synthetic milk | 1H-NMR | SIMC PLS | [153] |
| Beef | There is a need for new, non-invasive, rapid and reliable analytical methodologies that can easily be implemented and used for authentication of cattle production systems and the meat derived from them. Easily quantifiable markers could strengthen the current tracing methods for beef authentication. This study investigated the use of a nuclear magnetic resonance-based metabolomic approach as a tool to authenticate beef on the basis of the pre-slaughter production system | NMR | PCA PLS-DA | [154] |
| Cabbage | Cabbage (Brassica rapa ssp. pekinensis) is one of the most popular foods in Asia and is widely cultivated in many countries for the production of lightly fermented vegetables. In this study, metabolomic analysis was performed to distinguish two cultivars of cabbage grown in different geographical areas, Korea and China, using 1H-NMR spectroscopy coupled with multivariate statistical analysis. PCA showed clear discrimination between extracts of cabbage grown in Korea and China for two different cultivars (Chunmyeong and Chunjung) | NMR | PCA | [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emwas, A.-H.M.; Al-Rifai, N.; Szczepski, K.; Alsuhaymi, S.; Rayyan, S.; Almahasheer, H.; Jaremko, M.; Brennan, L.; Lachowicz, J.I. You Are What You Eat: Application of Metabolomics Approaches to Advance Nutrition Research. Foods 2021, 10, 1249. https://doi.org/10.3390/foods10061249
Emwas A-HM, Al-Rifai N, Szczepski K, Alsuhaymi S, Rayyan S, Almahasheer H, Jaremko M, Brennan L, Lachowicz JI. You Are What You Eat: Application of Metabolomics Approaches to Advance Nutrition Research. Foods. 2021; 10(6):1249. https://doi.org/10.3390/foods10061249
Chicago/Turabian StyleEmwas, Abdul-Hamid M., Nahla Al-Rifai, Kacper Szczepski, Shuruq Alsuhaymi, Saleh Rayyan, Hanan Almahasheer, Mariusz Jaremko, Lorraine Brennan, and Joanna Izabela Lachowicz. 2021. "You Are What You Eat: Application of Metabolomics Approaches to Advance Nutrition Research" Foods 10, no. 6: 1249. https://doi.org/10.3390/foods10061249