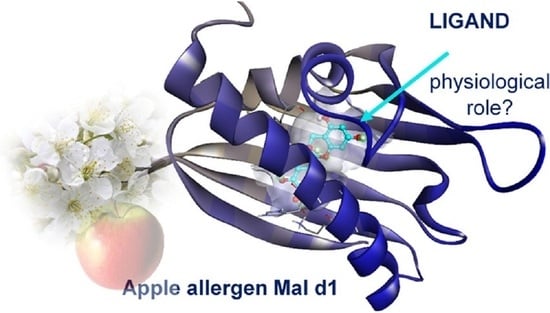
Microscale Thermophoresis Reveals Oxidized Glutathione as High-Affinity Ligand of Mal d 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Production and Purification of Recombinant rMal d 1
2.3. Microscale Thermophoresis
2.4. Molecular Docking
3. Results
3.1. rMal d 1.02 in Complex with Flavonoids
3.2. rMal d 1 Binds Glutathione in Both Reduced and Oxidized Forms with Different Specificities
3.3. Molecular Docking of the Identified Protein-Ligand Complexes
4. Discussion
4.1. Binding of Natural Flavonoids
4.2. Relationships of PR-10 Proteins, Glutathione, and Glutathione S-Transferase Expression
4.3. Effect of Ligand Binding on the Allergenicity of the Protein
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Vanek-Krebitz, M.; Hoffmann-Sommergruber, K.; Laimer Da Camara Machado, M.; Susani, M.; Ebner, C.; Kraft, D.; Scheiner, O.; Breiteneder, H. Cloning and sequencing of Mal d 1, the major allergen from apple (Malus domestica), and its immunological relationship to Bet v 1, the major birch pollen allergen. Biochem. Biophys. Res. Commun. 1995, 214, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Ausukua, M.; Dublin, I.; Echebarria, M.-A.; Aguirre, J.-M. Oral Allergy Syndrome (OAS). General and stomatological aspects. Med. Oral Patol. Oral Cir. Bucal 2009, 14, e568–e572. [Google Scholar] [CrossRef] [Green Version]
- Schimek, E.M.; Zwölfer, B.; Briza, P.; Jahn-Schmid, B.; Vogel, L.; Vieths, S.; Ebner, C.; Bohle, B. Gastrointestinal digestion of Bet v 1-homologous food allergens destroys their mediator-releasing, but not T cell-activating, capacity. J. Allergy Clin. Immunol. 2005, 116, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Bohle, B.; Zwölfer, B.; Heratizadeh, A.; Jahn-Schmid, B.; Antonia, Y.D.; Alter, M.; Keller, W.; Zuidmeer, L.; van Ree, R.; Werfel, T.; et al. Cooking birch pollen-related food: Divergent consequences for IgE- and T cell-mediated reactivity in vitro and in vivo. J. Allergy Clin. Immunol. 2006, 118, 242–249. [Google Scholar] [CrossRef]
- Costa, J.; Bavaro, S.L.; Benedé, S.; Diaz-Perales, A.; Bueno-Diaz, C.; Gelencser, E.; Klueber, J.; Larré, C.; Lozano-Ojalvo, D.; Lupi, R.; et al. Are Physicochemical Properties Shaping the Allergenic Potency of Plant Allergens? Clin. Rev. Allerg Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Hoffmann-Sommergruber, K. Plant Allergens and Pathogenesis-Related Proteins: What Do They Have in Common? Int. Arch. Allergy Immunol. 2000, 122, 155–166. [Google Scholar] [CrossRef]
- Schenk, M.F.; Cordewener, J.H.G.; America, A.H.P.; Van’t Westende, W.P.C.; Smulders, M.J.M.; Gilissen, L.J.W.J. Characterization of PR-10 genes from eight Betula species and detection of Bet v 1 isoforms in birch pollen. BMC Plant Biol. 2009, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajhede, M.; Osmark, P.; Poulsen, F.M.; Ipsen, H.; Larsen, J.N.; van Joost Neerven, R.J.; Schou, C.; Løwenstein, H.; Spangfort, M.D. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat. Struct. Biol. 1996, 3, 1040–1045. [Google Scholar] [CrossRef]
- Radauer, C.; Lackner, P.; Breiteneder, H. The Bet v 1 fold: An ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 2008, 8, 286. [Google Scholar] [CrossRef] [Green Version]
- Moiseyev, G.P.; Fedoreyeva, L.I.; Zhuravlev, Y.N.; Yasnetskaya, E.; Jekel, P.A.; Beintema, J.J. Primary structures of two ribonucleases from ginseng calluses. FEBS Lett. 1997, 407, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Besbes, F.; Franz-Oberdorf, K.; Schwab, W. Phosphorylation-dependent ribonuclease activity of Fra a 1 proteins. J. Plant Physiol. 2019, 233, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Casañal, A.; Zander, U.; Muñoz, C.; Dupeux, F.; Luque, I.; Botella, M.A.; Schwab, W.; Valpuesta, V.; Marquez, J.A. The strawberry pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J. Biol. Chem. 2013, 288, 35322–35332. [Google Scholar] [CrossRef] [Green Version]
- Seutter von Loetzen, C.; Hoffmann, T.; Hartl, M.J.; Schweimer, K.; Schwab, W.; Rösch, P.; Hartl-Spiegelhauer, O. Secret of the major birch pollen allergen Bet v 1: Identification of the physiological ligand. Biochem. J. 2014, 457, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Aglas, L.; Soh, W.T.; Kraiem, A.; Wenger, M.; Brandstetter, H.; Ferreira, F. Ligand Binding of PR-10 Proteins with a Particular Focus on the Bet v 1 Allergen Family. Curr. Allergy Asthma Rep. 2020, 20, 25. [Google Scholar] [CrossRef]
- Morris, J.S.; Caldo, K.M.P.; Liang, S.; Facchini, P.J. PR10/Bet v1-like Proteins as Novel Contributors to Plant Biochemical Diversity. Chembiochem 2021, 22, 264–287. [Google Scholar] [CrossRef]
- Casañal, A.; Zander, U.; Dupeux, F.; Valpuesta, V.; Marquez, J.A. Purification, crystallization and preliminary X-ray analysis of the strawberry allergens Fra a 1E and Fra a 3 in the presence of catechin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 510–514. [Google Scholar] [CrossRef] [Green Version]
- Neudecker, P.; Schweimer, K.; Nerkamp, J.; Scheurer, S.; Vieths, S.; Sticht, H.; Rösch, P. Allergic cross-reactivity made visible: Solution structure of the major cherry allergen Pru av 1. J. Biol. Chem. 2001, 276, 22756–22763. [Google Scholar] [CrossRef] [Green Version]
- Romanowski, M.J.; Soccio, R.E.; Breslow, J.L.; Burley, S.K. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc. Natl. Acad. Sci. USA 2002, 99, 6949–6954. [Google Scholar] [CrossRef] [Green Version]
- Kofler, S.; Asam, C.; Eckhard, U.; Wallner, M.; Ferreira, F.; Brandstetter, H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen bet v 1. J. Mol. Biol. 2012, 422, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Marković-Housley, Z.; Degano, M.; Lamba, D.; von Roepenack-Lahaye, E.; Clemens, S.; Susani, M.; Ferreira, F.; Scheiner, O.; Breiteneder, H. Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet v 1 and its Likely Biological Function as a Plant Steroid Carrier. J. Mol. Biol. 2003, 325, 123–133. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Ferreras, M.; Wimmer, R.; Petersen, S.V.; Enghild, J.J.; Otzen, D.E. The major allergen from birch tree pollen, Bet v 1, binds and permeabilizes membranes. Biochemistry 2007, 46, 3356–3365. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, J.E.; Wimmer, R.; Larsen, J.N.; Spangfort, M.D.; Otzen, D.E. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 2002, 277, 23684–23692. [Google Scholar] [CrossRef] [Green Version]
- Soh, W.T.; Aglas, L.; Mueller, G.A.; Gilles, S.; Weiss, R.; Scheiblhofer, S.; Huber, S.; Scheidt, T.; Thompson, P.M.; Briza, P.; et al. Multiple roles of Bet v 1 ligands in allergen stabilization and modulation of endosomal protease activity. Allergy 2019, 74, 2382–2393. [Google Scholar] [CrossRef] [Green Version]
- Seutter von Loetzen, C.; Jacob, T.; Hartl-Spiegelhauer, O.; Vogel, L.; Schiller, D.; Spörlein-Güttler, C.; Schobert, R.; Vieths, S.; Hartl, M.J.; Rösch, P. Ligand Recognition of the Major Birch Pollen Allergen Bet v 1 is Isoform Dependent. PLoS ONE 2015, 10, e0128677. [Google Scholar] [CrossRef] [Green Version]
- Jacob, T.; von Loetzen, C.S.; Reuter, A.; Lacher, U.; Schiller, D.; Schobert, R.; Mahler, V.; Vieths, S.; Rösch, P.; Schweimer, K.; et al. Identification of a natural ligand of the hazel allergen Cor a 1. Sci. Rep. 2019, 9, 8714. [Google Scholar] [CrossRef] [PubMed]
- Zubini, P.; Zambelli, B.; Musiani, F.; Ciurli, S.; Bertolini, P.; Baraldi, E. The RNA hydrolysis and the cytokinin binding activities of PR-10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol. 2009, 150, 1235–1247. [Google Scholar] [CrossRef] [Green Version]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Romer, E.; Chebib, S.; Bergmann, K.-C.; Plate, K.; Becker, S.; Ludwig, C.; Meng, C.; Fischer, T.; Dierend, W.; Schwab, W. Tiered approach for the identification of Mal d 1 reduced, well tolerated apple genotypes. Sci. Rep. 2020, 10, 9144. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Gill, C.S.; von Hippel, P.H. Calculation of Protein Extinction Coefficients from Amino Acid Squence Data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlovskii, I.; Popov, P. Protein-Peptide Binding Site Detection Using 3D Convolutional Neural Networks. J. Chem. Inf. Model. 2021, 61, 3814–3823. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Salces, R.M.; Ndjoko, K.; Queiroz, E.F.; Ioset, J.R.; Hostettmann, K.; Berrueta, L.A.; Gallo, B.; Vicente, F. On-line characterisation of apple polyphenols by liquid chromatography coupled with mass spectrometry and ultraviolet absorbance detection. J. Chromatogr. A 2004, 1046, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Ban, Z.; Jiang, Y. Variation in Antioxidant Metabolites and Enzymes of ‘Red Fuji’ Apple Pulp and Peel During Cold Storage. Int. J. Food Prop. 2014, 17, 1067–1080. [Google Scholar] [CrossRef]
- Ahammer, L.; Grutsch, S.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Structure of the Major Apple Allergen Mal d 1. J. Agric. Food Chem. 2017, 65, 1606–1612. [Google Scholar] [CrossRef]
- Jakobek, L.; García-Villalba, R.; Tomás-Barberán, F.A. Polyphenolic characterisation of old local apple varieties from Southeastern European region. J. Food Compost Anal. 2013, 31, 199–211. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [Green Version]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Treutter, D. Biosynthesis of phenolic compounds and its regulation in apple. Plant Growth Regul. 2001, 34, 71–89. [Google Scholar] [CrossRef]
- Muñoz, C.; Hoffmann, T.; Escobar, N.M.; Ludemann, F.; Botella, M.A.; Valpuesta, V.; Schwab, W. The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol. Plant 2010, 3, 113–124. [Google Scholar] [CrossRef]
- Meister, A. Glutathione metabolism and its selective modification. J. Biol. Chem. 1988, 263, 17205–17208. [Google Scholar] [CrossRef]
- Ketterer, B.; Coles, B.; Meyer, D.J. The role of glutathione in detoxication. Environ. Health Perspect. 1983, 49, 59–69. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Li, Y.; Di, B.; Zhang, J.; Liu, Y. Effect of high temperature and excessive light on glutathione content in apple peel. Front. Agric. China 2008, 2, 97–102. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Ko, K.; Chang, W.-L.; Kuo, W.-C.; Chen, G.-H.; Lin, T.-P. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015, 83, 926–939. [Google Scholar] [CrossRef]
- Koistinen, K.M.; Soininen, P.; Venäläinen, T.A.; Häyrinen, J.; Laatikainen, R.; Peräkylä, M.; Tervahauta, A.I.; Kärenlampi, S.O. Birch PR-10c interacts with several biologically important ligands. Phytochemistry 2005, 66, 2524–2533. [Google Scholar] [CrossRef]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.; Dean, J.V. Transport of Anthocyanins and other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Sci Rep. 2019, 9, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingate, V.P.; Lawton, M.A.; Lamb, C.J. Glutathione causes a massive and selective induction of plant defense genes. Plant Physiol. 1988, 87, 206–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hjernø, K.; Alm, R.; Canbäck, B.; Matthiesen, R.; Trajkovski, K.; Björk, L.; Roepstorff, P.; Emanuelsson, C. Down-regulation of the strawberry Bet v 1-homologous allergen in concert with the flavonoid biosynthesis pathway in colorless strawberry mutant. Proteomics 2006, 6, 1574–1587. [Google Scholar] [CrossRef]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3, REVIEWS3004. [Google Scholar] [CrossRef] [Green Version]
- Strange, R.C.; Spiteri, M.A.; Ramachandran, S.; Fryer, A.A. Glutathione-S-transferase family of enzymes. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2001, 482, 21–26. [Google Scholar] [CrossRef]
- Härtl, K.; Denton, A.; Franz-Oberdorf, K.; Hoffmann, T.; Spornraft, M.; Usadel, B.; Schwab, W. Early metabolic and transcriptional variations in fruit of natural white-fruited Fragaria vesca genotypes. Sci. Rep. 2017, 7, 45113. [Google Scholar] [CrossRef] [Green Version]
- Goodman, C.D.; Casati, P.; Walbot, V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef] [Green Version]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. EXBOTJ 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis vinífera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orozco-Navarrete, B.; Song, J.; Casañal, A.; Sozzani, R.; Flors, V.; Sánchez-Sevilla, J.F.; Trinkl, J.; Hoffmann, T.; Merchante, C.; Schwab, W.; et al. Down-regulation of Fra a 1.02 in strawberry fruits causes transcriptomic and metabolic changes compatible with an altered defense response. Hortic. Res. 2021, 8, 58. [Google Scholar] [CrossRef]
- Marzban, G.; Puehringer, H.; Dey, R.; Brynda, S.; Ma, Y.; Martinelli, A.; Zaccarini, M.; van der Weg, E.; Housley, Z.; Kolarich, D.; et al. Localisation and distribution of the major allergens in apple fruits. Plant Sci. 2005, 169, 387–394. [Google Scholar] [CrossRef]
- Breiteneder, H.; Mills, E.N.C. Molecular properties of food allergens. J. Allergy Clin. Immunol. 2005, 115, 14–23. [Google Scholar] [CrossRef]
- Machado, Y.; Freier, R.; Scheiblhofer, S.; Thalhamer, T.; Mayr, M.; Briza, P.; Grutsch, S.; Ahammer, L.; Fuchs, J.E.; Wallnoefer, H.G.; et al. Fold stability during endolysosomal acidification is a key factor for allergenicity and immunogenicity of the major birch pollen allergen. J. Allergy Clin. Immunol. 2016, 137, 1525–1534. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; van de Weg, E.W.; Matos, C.I.; Arens, P.; Bolhaar, S.T.H.P.; Knulst, A.C.; Li, Y.; Hoffmann-Sommergruber, K.; Gilissen, L.J.W.J. Assessment of allelic diversity in intron-containing Mal d 1 genes and their association to apple allergenicity. BMC Plant Biol. 2008, 8, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliarani, G.; Paris, R.; Iorio, A.R.; Tartarini, S.; Del Duca, S.; Arens, P.; Peters, S.; van de Weg, E. Genomic organisation of the Mal d 1 gene cluster on linkage group 16 in apple. Mol. Breed. 2012, 29, 759–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spangfort, M.D.; Mirza, O.; Ipsen, H.; van Neerven, R.J.J.; Gajhede, M.; Larsen, J.N. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J. Immunol. 2003, 171, 3084–3090. [Google Scholar] [CrossRef] [Green Version]
- Son, D.Y.; Scheurer, S.; Hoffmann, A.; Haustein, D.; Vieths, S. Pollen-related food allergy: Cloning and immunological analysis of isoforms and mutants of Mal d 1, the major apple allergen, and Bet v 1, the major birch pollen allergen. Eur. J. Nutr. 1999, 38, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Gadermaier, G.; Bohle, B.; Bolhaar, S.; Knulst, A.; Markovic-Housley, Z.; Breiteneder, H.; Briza, P.; Hoffmann-Sommergruber, K.; Ferreira, F. Mutational analysis of amino acid positions crucial for IgE-binding epitopes of the major apple (Malus domestica) allergen, Mal d 1. Int. Arch. Allergy Immunol. 2006, 139, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Eidelpes, R.; Hofer, F.; Röck, M.; Führer, S.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Structure and Zeatin Binding of the Peach Allergen Pru p 1. J. Agric. Food Chem. 2021, 69, 8120–8129. [Google Scholar] [CrossRef] [PubMed]
- Asam, C.; Batista, A.L.; Moraes, A.H.; de Paula, V.S.; Almeida, F.C.L.; Aglas, L.; Kitzmüller, C.; Bohle, B.; Ebner, C.; Ferreira, F.; et al. Bet v 1--a Trojan horse for small ligands boosting allergic sensitization? Clin. Exp. Allergy 2014, 44, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Grutsch, S.; Fuchs, J.E.; Freier, R.; Kofler, S.; Bibi, M.; Asam, C.; Wallner, M.; Ferreira, F.; Brandstetter, H.; Liedl, K.R.; et al. Ligand binding modulates the structural dynamics and compactness of the major birch pollen allergen. Biophys. J. 2014, 107, 2972–2981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, M.; Davies, A.M.; Liljekvist, M.; Carlsson, F.; Gould, H.J.; Sutton, B.J.; Ohlin, M. Human IgE against the major allergen Bet v 1--defining an epitope with limited cross-reactivity between different PR-10 family proteins. Clin. Exp. Allergy 2014, 44, 288–299. [Google Scholar] [CrossRef]
- Führer, S.; Kamenik, A.S.; Zeindl, R.; Nothegger, B.; Hofer, F.; Reider, N.; Liedl, K.R.; Tollinger, M. Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens. Sci. Rep. 2021, 11, 4173. [Google Scholar] [CrossRef]
- Grutsch, S.; Fuchs, J.E.; Ahammer, L.; Kamenik, A.S.; Liedl, K.R.; Tollinger, M. Conformational Flexibility Differentiates Naturally Occurring Bet v 1 Isoforms. Int. J. Mol. Sci. 2017, 18, 1192. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.; Radauer, C.; Bublin, M.; Hoffmann-Sommergruber, K.; Kopp, T.; Greisenegger, E.K.; Vogel, L.; Vieths, S.; Scheiner, O.; Breiteneder, H. Naturally occurring hypoallergenic Bet v 1 isoforms fail to induce IgE responses in individuals with birch pollen allergy. J. Allergy Clin. Immunol. 2008, 121, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Smole, U.; Balazs, N.; Hoffmann-Sommergruber, K.; Radauer, C.; Hafner, C.; Wallner, M.; Ferreira, F.; Grössinger, R.; de Jong, E.C.; Wagner, S.; et al. Differential T-cell responses and allergen uptake after exposure of dendritic cells to the birch pollen allergens Bet v 1.0101, Bet v 1.0401 and Bet v 1.1001. Immunobiology 2010, 215, 903–909. [Google Scholar] [CrossRef]




| Plant | Protein | Ligand | Kd [µM] | Method | References |
|---|---|---|---|---|---|
| Birch | Bet v 1a | ANS | 18.5 | NMR, Fluorescence | [20,23] |
| Deoxycholate | 58.8 | NMR, SAW | [20,24] | ||
| Quercetin | 31.4 | NMR | [14,25] | ||
| 9.2 | UV/VIS | [14,25] | |||
| Quercetin-3-O-sophoroside | 0.57 | Fluorescence | [14,25] | ||
| Quercetin-3-O-galactoside | <5 | NMR | [25] | ||
| Quercetin-3-O-glucoside | 288.4 | NMR | [25] | ||
| Fisetin | 14.3 | UV/VIS | [25] | ||
| Myricetin | 14.6 | NMR | [25] | ||
| Naringenin | 60.6 | UV/VIS | [25] | ||
| Bet v 1m | Quercetin | 65.8 | NMR | [25] | |
| 26.5 | UV/VIS | [25] | |||
| Quercetin-3-O-galactoside | <5 | NMR | [25] | ||
| Quercetin-3-O-glucoside | <5 | NMR | [25] | ||
| Fisetin | 68.6 | UV/VIS | [25] | ||
| Myricetin | 99.3 | NMR | [25] | ||
| Naringenin | 28.1 | UV/VIS | [25] | ||
| Bet v 1d | Quercetin | 10.2 | UV/VIS | [25] | |
| Fisetin | 13.9 | UV/VIS | [25] | ||
| Myricetin | 1.2 | UV/VIS | [25] | ||
| Naringenin | 37.7 | UV/VIS | [25] | ||
| Strawberry | Fra a 1E | Quercetin-3-O-glucuronide | 5.3 | ITC | [13] |
| Fra a 2 | Myricetin | 19.5 | ITC | [13] | |
| Fra a 3 | (+)-Catechin | 8.9 | ITC | [13] | |
| Hazelnut | Cor a 1 | Quercetin-3-O-(2-O-β-D-glucopyranosyl)-β-D-galactopyranoside | <5 | NMR | [26] |
| Peach | Pru p 1.0101 | Zeatin | 9.4 | ITC | [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chebib, S.; Schwab, W. Microscale Thermophoresis Reveals Oxidized Glutathione as High-Affinity Ligand of Mal d 1. Foods 2021, 10, 2771. https://doi.org/10.3390/foods10112771
Chebib S, Schwab W. Microscale Thermophoresis Reveals Oxidized Glutathione as High-Affinity Ligand of Mal d 1. Foods. 2021; 10(11):2771. https://doi.org/10.3390/foods10112771
Chicago/Turabian StyleChebib, Soraya, and Wilfried Schwab. 2021. "Microscale Thermophoresis Reveals Oxidized Glutathione as High-Affinity Ligand of Mal d 1" Foods 10, no. 11: 2771. https://doi.org/10.3390/foods10112771