Synergistic Effects of Limosilactobacillus fermentum ASBT-2 with Oxyresveratrol Isolated from Coconut Shell Waste
Abstract
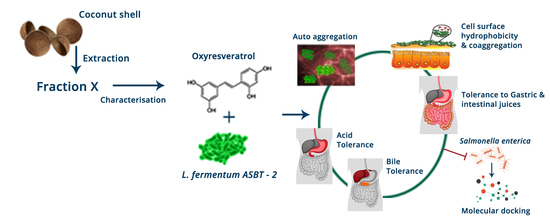
:1. Introduction
2. Materials and Methods
2.1. Raw Materials; Chemicals and Bacterial Strains
2.2. Extraction and Isolation of Oxyresveratrol
Characterization of Oxyresveratrol
2.3. Determination of Minimum Inhibitory Concentration (MIC) of Oxyresveratrol
2.4. Effect of Oxyresveratrol on pH Tolerance of the Strain
2.5. Effect of Oxyresveratrol on Tolerance to Bile Salts
2.6. Effects on Survival under Simulated Gastrointestinal Conditions
2.7. Effect of Oxyresveratrol on Cell Surface Hydrophobicity of the Strain
2.8. Effects on Autoaggregation Potential of the Strain
2.9. Effects of Oxyresveratrol on Coaggregation Potential
2.10. Effects of Oxyresveratrol on Antagonistic Activity of the Strain
2.11. Statistical Analysis
2.12. GenBank Accession Numbers
2.13. Computational Molecular Docking Studies
3. Results
3.1. Bacterial Strains and Raw Materials
3.2. Extraction and Isolation of Oxyresveratrol
Characterization of Oxyresveratrol
3.3. Determination of Minimum Inhibitory Concentration (MIC) of Oxyresveratrol
3.4. Effect of Oxyresveratrol on pH Tolerance of the Strain
3.5. Effect of Oxyresveratrol on Tolerance to Bile Salts
3.6. Effects on Survival under Simulated Gastrointestinal Conditions
3.7. Effect of Oxyresveratrol on Cell Surface Hydrophobicity
3.8. Effects on Autoaggregation Potential of the Strain
3.9. Effects of Oxyresveratrol on Coaggregation Potential
3.10. Effects of Oxyresveratrol on Antagonistic Activity of the Strain
3.11. Molecular Docking Study Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antonio, A.L.; Pereira, E.; Pinela, J.; Heleno, S.; Pereira, C.; Ferreira, I.C. Determination of Antioxidant Compounds in Foodstuff. In Food Safety: Innovative Analytical Tools for Safety Assessment; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2016; pp. 179–220. [Google Scholar] [CrossRef] [Green Version]
- Mojzer, E.B.; Hrnčič, M.K.; Škerget, M.; Knez Hrnčič, M.; Bren, U. Polyphenols: Extraction Methods, Antioxidative Action, Bioavailability and Anticarcinogenic Effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef]
- Rasouli, H.; Farzaei, M.H.; Khodarahmi, R. Polyphenols and their benefits: A review. Int. J. Food Prop. 2017, 20, 1700–1742. [Google Scholar] [CrossRef] [Green Version]
- Paolillo, R.; Carratelli, C.R.; Rizzo, A. Effect of resveratrol and quercetin in experimental infection by Salmonella enterica serovar typhimurium. Int. Immunopharmacol. 2011, 11, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Mattio, L.; Catinella, G.; Dallavalle, S.; Pinto, A. Stilbenoids: A Natural Arsenal against Bacterial Pathogens. Antibiotics 2020, 9, 336. [Google Scholar] [CrossRef]
- Santos, A.; De Albuquerque, T.M.R.; Alves, J.L.D.B.; De Souza, E.L. Effects of Quercetin and Resveratrol on in vitro Properties Related to the Functionality of Potentially Probiotic Lactobacillus Strains. Front. Microbiol. 2019, 10, 2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Chao, L.; Wei, X.; Hu, C.; XiaoLi, Q.; XianZhi, H. Advance in the Study of Oxyreseveratrol. Int. J. Pharmacol. 2014, 10, 44–54. [Google Scholar]
- DebMandal, M.; Mandal, S. Coconut (Cocos nucifera L.: Arecaceae): In health promotion and disease prevention. Asian Pac. J. Trop. Med. 2011, 4, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Tarawan, V.M.; Mantilidewi, K.I.; Dhini, I.M.; Radhiyanti, P.T.; Sutedja, E. Coconut Shell Liquid Smoke Promotes Burn Wound Healing. J. Evid. Based Integr. Med. 2016, 22, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.S.; Bose, C.; Shaji, S.K.; Pandurangan, N.; Kumar, G.B.; Nair, B.G.; Banerji, A. Coconut shell derived bioactive compound Oxyresveratrol mediates regulation of Matrix metalloproteinase 9. Int. J. Pharma Bio Sci. 2017, 8, 202–210. [Google Scholar] [CrossRef]
- Cueva, C.; Gil-Sánchez, I.; Ayuda-Durán, B.; González-Manzano, S.; González-Paramás, A.M.; Santos-Buelga, C.; Bartolomé, B.; Moreno-Arribas, M.V. An Integrated View of the Effects of Wine Polyphenols and Their Relevant Metabolites on Gut and Host Health. Molecules 2017, 22, 99. [Google Scholar] [CrossRef]
- Sharma, R.; Padwad, Y. Plant-polyphenols based second-generation synbiotics: Emerging concepts, challenges, and opportunities. Nutrition 2020, 77, 110785. [Google Scholar] [CrossRef]
- Yin, R.; Kuo, H.-C.; Hudlikar, R.; Sargsyan, D.; Li, S.; Wang, L.; Wu, R.; Kong, A.-N. Gut Microbiota, Dietary Phytochemicals, and Benefits to Human Health. Curr. Pharmacol. Rep. 2019, 5, 332–344. [Google Scholar] [CrossRef]
- Kemperman, R.A.; Bolca, S.; Roger, L.C.; Vaughan, E.E. Novel approaches for analysing gut microbes and dietary polyphenols: Challenges and opportunities. Microbiology 2010, 156, 3224–3231. [Google Scholar] [CrossRef] [Green Version]
- Babu, P.; Veedu, A.P.; Prakash, V.; Prasad, M.; Salim, A.; Madhavan, A.; Nair, B.G.; Pal, S. Draft Genome Sequence of a Lactobacillus fermentum Strain Isolated from Domestic Sewage in Kerala, India. Microbiol. Resour. Announc. 2020, 9, e00713-20. [Google Scholar] [CrossRef]
- Mena, P.; Cirlini, M.; Tassotti, M.; Herrlinger, K.A.; Dall’Asta, C.; Del Rio, D. Phytochemical Profiling of Flavonoids, Phenolic Acids, Terpenoids, and Volatile Fraction of a Rosemary (Rosmarinus officinalis L.) Extract. Molecules 2016, 21, 1576. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Zhang, X.; Li, S.; Li, C.; Li, D.; Yang, Z. Evaluation of probiotic properties of Lactobacillus plantarum strains isolated from Chinese sauerkraut. World J. Microbiol. Biotechnol. 2012, 29, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Prabhurajeshwar, C.; Chandrakanth, K. Evaluation of antimicrobial properties and their substances against pathogenic bacteria in-vitro by probiotic Lactobacilli strains isolated from commercial yoghurt. Clin. Nutr. Exp. 2019, 23, 97–115. [Google Scholar] [CrossRef] [Green Version]
- Todorov, S.; Botes, M.; Guigas, C.; Schillinger, U.; Wiid, I.; Wachsman, M.; Holzapfel, W.; Dicks, L. Boza, a natural source of probiotic lactic acid bacteria. J. Appl. Microbiol. 2007, 104, 465–477. [Google Scholar] [CrossRef]
- Agaliya, P.J. Screening of Lactobacillus plantarum isolated from fermented idli batter for probiotic properties. Afr. J. Biotechnol. 2012, 11, 12856–12864. [Google Scholar] [CrossRef]
- Berkes, E.; Liao, Y.-H.; Neef, D.; Grandalski, M.; Monsul, N. Potentiated In Vitro Probiotic Activities of Lactobacillus fermentum LfQi6 Biofilm Biomass Versus Planktonic Culture. Probiotics Antimicrob. Proteins 2019, 12, 1097–1114. [Google Scholar] [CrossRef]
- Klein, S. The Use of Biorelevant Dissolution Media to Forecast the In Vivo Performance of a Drug. AAPS J. 2010, 12, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; de Melo Franco, B.D. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 2007, 226, 1065–1073. [Google Scholar] [CrossRef]
- Halder, D.; Mandal, M.; Chatterjee, S.S.; Pal, N.K.; Mandal, S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines 2017, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Binkowski, T.A. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003, 31, 3352–3355. [Google Scholar] [CrossRef] [Green Version]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A Fast Flexible Docking Method using an Incremental Construction Algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef] [Green Version]
- Schneider, N.; Lange, G.; Hindle, S.; Klein, R.; Rarey, M. A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: Methods behind the HYDE scoring function. J. Comput. Mol. Des. 2012, 27, 15–29. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Kim, J.-H.; Kim, K.-J. Crystal Structure of the Salmonella enterica Serovar typhimurium Virulence Factor SrfJ, a Glycoside Hydrolase Family Enzyme. J. Bacteriol. 2009, 191, 6550–6554. [Google Scholar] [CrossRef] [Green Version]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6, 116. [Google Scholar] [CrossRef] [Green Version]
- Alves-Santos, A.M.; Sugizaki, C.S.A.; Lima, G.C.; Naves, M.M.V. Prebiotic effect of dietary polyphenols: A systematic review. J. Funct. Foods 2020, 74, 104169. [Google Scholar] [CrossRef]
- Thilakarathna, W.W.; Langille, M.G.; Rupasinghe, H.V. Polyphenol-based prebiotics and synbiotics: Potential for cancer chemoprevention. Curr. Opin. Food Sci. 2018, 20, 51–57. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Varma, P.; Nisha, N.; Dinesh, K.R.; Kumar, A.V.; Biswas, R. Anti-Infective Properties of Lactobacillus fermentum against Staphylococcus aureus and Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 2011, 20, 137–143. [Google Scholar] [CrossRef]
- Hervert-Hernández, D.; Pintado, C.; Rotger, R.; Goñi, I. Stimulatory role of grape pomace polyphenols on Lactobacillus acidophilus growth. Int. J. Food Microbiol. 2009, 136, 119–122. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Andrian, E.; Grenier, D. Resveratrol attenuates the pathogenic and inflammatory properties of Porphyromonas gingivalis. Mol. Oral Microbiol. 2019, 34, 118–130. [Google Scholar] [CrossRef]
- O’Connor, D.J.; Wong, R.W.K.; Rabie, A.B.M. Resveratrol Inhibits Periodontal Pathogens In Vitro. Phytother. Res. 2011, 25, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The Reciprocal Interactions between Polyphenols and Gut Microbiota and Effects on Bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef]
- Singh, A.K.; Cabral, C.; Kumar, R.; Ganguly, R.; Rana, H.K.; Gupta, A.; Lauro, M.R.; Carbone, C.; Reis, F.; Pandey, A.K. Beneficial Effects of Dietary Polyphenols on Gut Microbiota and Strategies to Improve Delivery Efficiency. Nutrients 2019, 11, 2216. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligne, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shokryazdan, P.; Jahromi, M.F.; Liang, J.B.; Ho, Y.W. Probiotics: From Isolation to Application. J. Am. Coll. Nutr. 2017, 36, 666–676. [Google Scholar] [CrossRef]
- Chambers, K.F.; Day, P.E.; Aboufarrag, H.T.; Kroon, P.A. Polyphenol Effects on Cholesterol Metabolism via Bile Acid Biosynthesis, CYP7A1: A Review. Nutrients 2019, 11, 2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mountzouris, K.C.; Kotzampassi, K.; Tsirtsikos, P.; Kapoutzis, K.; Fegeros, K. Effects of Lactobacillus acidophilus on gut microflora metabolic biomarkers in fed and fasted rats. Clin. Nutr. 2009, 28, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [Green Version]
- Volstatova, T.; Marsik, P.; Rada, V.; Geigerova, M.; Havlik, J. Effect of apple extracts and selective polyphenols on the adhesion of potential probiotic strains of Lactobacillus gasseri R and Lactobacillus casei FMP. J. Funct. Foods 2017, 35, 391–397. [Google Scholar] [CrossRef]
- Bustos, I.; García-Cayuela, T.; Hernández-Ledesma, B.; Peláez, C.; Requena, T.; Martínez-Cuesta, M.C. Effect of Flavan-3-ols on the Adhesion of Potential Probiotic Lactobacilli to Intestinal Cells. J. Agric. Food Chem. 2012, 60, 9082–9088. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Kos, B.; Šušković, J.; Vuković, S.; Šimpraga, M.; Frece, J.; Matošić, S. Adhesion and aggregation ability of probiotic strainLactobacillus acidophilusM92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Likhitwitayawuid, K. Oxyresveratrol: Sources, Productions, Biological Activities, Pharmacokinetics, and Delivery Systems. Molecules 2021, 26, 4212. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Raorane, C.J.; Ryu, S.Y.; Shim, J.-J.; Lee, J. The anti-biofilm and anti-virulence activities of trans-resveratrol and oxyresveratrol against uropathogenic Escherichia coli. Biofouling 2019, 35, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.-Y.; Chen, T.-T.; Tan, X.-J.; Chen, T.; Jia, A.-Q. The quorum-sensing inhibiting effects of stilbenoids and their potential structure–activity relationship. Bioorganic Med. Chem. Lett. 2015, 25, 5217–5220. [Google Scholar] [CrossRef]
- Joung, D.-K.; Choi, S.-H.; Kang, O.-H.; Kim, S.-B.; Mun, S.-H.; Seo, Y.-S.; Kang, D.-H.; Gong, R.; Shin, D.-W.; Kim, Y.-C.; et al. Synergistic effects of oxyresveratrol in conjunction with antibiotics against methicillin-resistant Staphylococcus aureus. Mol. Med. Rep. 2012, 12, 663–667. [Google Scholar] [CrossRef]
- Huang, H.-L.; Zhang, J.-Q.; Chen, G.-T.; Lu, Z.-Q.; Sha, N.; Guo, D.-A. Simultaneous Determination of Oxyresveratrol and Resveratrol in Rat Bile and Urine by HPLC after Oral Administration of Smilax china Extract. Nat. Prod. Commun. 2009, 4, 1934578X0900400617. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-C.; Tseng, C.-H.; Wang, P.-W.; Lu, P.-L.; Weng, Y.-H.; Yen, F.-L.; Fang, J.-Y. Pterostilbene, a Methoxylated Resveratrol Derivative, Efficiently Eradicates Planktonic, Biofilm, and Intracellular MRSA by Topical Application. Front. Microbiol. 2017, 8, 1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero-Alba, M.; Bernal-Bayard, J.; Ramos-Morales, F. SrfJ, a Salmonella Type III Secretion System Effector Regulated by PhoP, RcsB, and IolR. J. Bacteriol. 2012, 194, 4226–4236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilera-Herce, J.; Zarkani, A.A.; Schikora, A.; Ramos-Morales, F. Dual Expression of the Salmonella Effector SrfJ in Mammalian Cells and Plants. Front. Microbiol. 2017, 8, 2410. [Google Scholar] [CrossRef] [PubMed]
- Kores, K.; Lešnik, S.; Bren, U.; Janežič, D.; Konc, J. Discovery of Novel Potential Human Targets of Resveratrol by Inverse Molecular Docking. J. Chem. Inf. Model. 2019, 59, 2467–2478. [Google Scholar] [CrossRef] [PubMed]



















| Sl.No. | Compound | Binding Affinity | Ligand Efficiency | LogP | Torsions | Clashes |
|---|---|---|---|---|---|---|
| 1 | Oxyresveratrol | μM-mM | Acceptable | 2.97 | Matches with CSD | No Clashes |
| 2 | Resveratrol | Towards mM | Not Acceptable | 2.05 | Matches with CSD | Inter Clashes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prakash, V.; Krishnan, A.S.; Ramesh, R.; Bose, C.; Pillai, G.G.; Nair, B.G.; Pal, S. Synergistic Effects of Limosilactobacillus fermentum ASBT-2 with Oxyresveratrol Isolated from Coconut Shell Waste. Foods 2021, 10, 2548. https://doi.org/10.3390/foods10112548
Prakash V, Krishnan AS, Ramesh R, Bose C, Pillai GG, Nair BG, Pal S. Synergistic Effects of Limosilactobacillus fermentum ASBT-2 with Oxyresveratrol Isolated from Coconut Shell Waste. Foods. 2021; 10(11):2548. https://doi.org/10.3390/foods10112548
Chicago/Turabian StylePrakash, Vidhya, Akshaya S Krishnan, Reshma Ramesh, Chinchu Bose, Girinath G. Pillai, Bipin G. Nair, and Sanjay Pal. 2021. "Synergistic Effects of Limosilactobacillus fermentum ASBT-2 with Oxyresveratrol Isolated from Coconut Shell Waste" Foods 10, no. 11: 2548. https://doi.org/10.3390/foods10112548