Bio-Guided Fractionation of Ethanol Extract of Leaves of Esenbeckia alata Kunt (Rutaceae) Led to the Isolation of Two Cytotoxic Quinoline Alkaloids: Evidence of Selectivity Against Leukemia Cells
Abstract
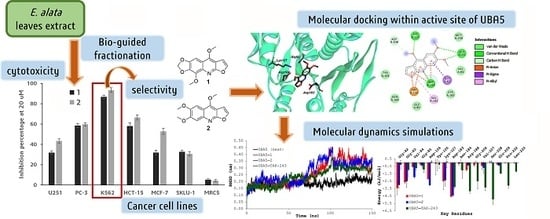
:1. Introduction
2. Experimental Section
2.1. Plant Material
2.2. Bio-Guided Fractionation
2.3. Cytotoxic Activity Test
2.4. Molecular Docking
2.5. Molecular Dynamics Simulations
2.6. Binding Free Energy Calculation
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yu, X.; Sun, S.; Guo, Y.; Liu, Y.; Yang, D.; Li, G.; Lü, S. Citri Reticulatae Pericarpium (Chenpi): Botany, ethnopharmacology, phytochemistry, and pharmacology of a frequently used traditional Chinese medicine. J. Ethnopharmacol. 2018, 220, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Pollio, A.; De Natale, A.; Appetiti, E.; Aliotta, G.; Touwaide, A. Continuity and change in the Mediterranean medical tradition: Ruta spp. (rutaceae) in Hippocratic medicine and present practices. J. Ethnopharmacol. 2008, 116, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Groppo, M.; Pirani, J.R.; Salatino, M.L.F.; Blanco, S.R.; Kallunki, J.A. Phylogeny of Rutaceae based on twononcoding regions from cpDNA. Am. J. Bot. 2008, 95, 985–1005. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Lopes, E.M.; Maier, J.A.; da Silva, M.R.; Regasini, L.O.; Simote, S.Y.; Lopes, N.P.; Pirani, J.R.; da Silva Bolzani, V.; Young, M.C.M. Alkaloids from stems of Esenbeckia leiocarpa Engl. (Rutaceae) as potential treatment for Alzheimer disease. Molecules 2010, 15, 9205–9213. [Google Scholar] [CrossRef] [PubMed]
- Mata, R.; Macías, M.L.; Rojas, I.S.; Lotina-Hennsen, B.; Toscano, R.A.; Anaya, A.L. Phytotoxic compounds from Esenbeckia yaxhoob. Phytochemistry 1998, 49, 441–449. [Google Scholar] [CrossRef]
- Anaya, A.L.; Macías-Rubalcava, M.; Cruz-Ortega, R.; García-Santana, C.; Sánchez-Monterrubio, P.N.; Hernández-Bautista, B.E.; Mata, R. Allelochemicals from Stauranthus perforatus, a Rutaceous tree of the Yucatan Peninsula, Mexico. Phytochemistry 2005, 66, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, D.L. Alkaloids, limonoids and furocoumarins from three Mexican Esenbeckia species. Phytochemistry 1980, 19, 941–944. [Google Scholar] [CrossRef]
- Rios, M.Y.; Delgado, G. Polyprenols and acylphloroglucinols from Esenbeckia nesiotica. Phytochemistry 1992, 31, 3491–3494. [Google Scholar] [CrossRef]
- García Beltrán, O.; Cuca Suárez, L.E. Constituyentes no polares de la corteza de Esenbeckia alata y actividad antimicrobiana. Rev. Colomb. Quim. 2003, 32, 23–28. [Google Scholar]
- Cuca Suárez, L.E.; Coy Barrera, C.A. Metabolites isolated from Esenbeckia alata (Karst & Triana) (Rutaceae). Biochem. Syst. Ecol. 2007, 35, 386–388. [Google Scholar]
- Cuca-Suarez, L.E.; Coy-Barrera, E.D.; Alvarez-Caballero, J.M. Quinoline alkaloids and friedelane-type triterpenes isolated from leaves and wood of Esenbeckia alata kunt (Rutaceae). Quim. Nova 2011, 34, 984–986. [Google Scholar] [CrossRef]
- Trani, M.; Carbonetti, A.; Delle Monache, G.; Delle Monache, F. Dihydrochalcones and coumarins of Esenbeckia grandiflora subsp. brevipetiolata. Fitoterapia 2004, 75, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Rios, M.Y.; Rosas-Alonso, E.; Berenice Aguilar-Guadarrama, A. Alkaloids, coumarins and sesquiterpenes from Esenbeckia conspecta Kunt (Rutaceae). Biochem. Syst. Ecol. 2002, 30, 367–369. [Google Scholar] [CrossRef]
- Guilhon, G.M.S.P.; Baetas, A.C.S.; Maia, J.G.S.; Conserva, L.M. 2-Alkyl-4-quinolone alkaloids and cinnamic acid derivatives from Esenbeckia almawillia. Phytochemistry 1994, 37, 1193–1195. [Google Scholar] [CrossRef]
- Simpson, D.S.; Jacobs, H. Alkaloids and coumarins from Esenbeckia pentaphylla (Rutaceae). Biochem. Syst. Ecol. 2005, 33, 841–844. [Google Scholar] [CrossRef]
- Shang, X.-F.; Morris-Natschke, S.L.; Liu, Y.-Q.; Guo, X.; Xu, X.-S.; Goto, M.; Li, J.-C.; Yang, G.-Z.; Lee, K.-H. Biologically active quinoline and quinazoline alkaloids part I. Med. Res. Rev. 2018, 38, 775–828. [Google Scholar] [CrossRef]
- Byler, K.G.; Wang, C.; Setzer, W.N. Quinoline alkaloids as intercalative topoisomerase inhibitors. J. Mol. Model. 2009, 15, 1417–1426. [Google Scholar] [CrossRef]
- Kuete, V.; Fouotsa, H.; Mbaveng, A.T.; Wiench, B.; Nkengfack, A.E.; Efferth, T. Cytotoxicity of a naturally occurring furoquinoline alkaloid and four acridone alkaloids towards multi-factorial drug-resistant cancer cells. Phytomedicine 2015, 22, 946–951. [Google Scholar] [CrossRef]
- Mohammed, M.M.D.; El-Sharkawy, E.R. Cytotoxic new furoquinoline alkaloid isolated from Ammi majus L. growing in Egypt. Nat. Prod. Res. 2017, 31, 645–652. [Google Scholar] [CrossRef]
- Xu, G.W.; Ali, M.; Wood, T.E.; Wong, D.; Maclean, N.; Wang, X.; Gronda, M.; Skrtic, M.; Li, X.; Hurren, R.; et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood 2010, 115, 2251–2259. [Google Scholar] [CrossRef] [Green Version]
- Sahasrabuddhe, A.A.; Elenitoba-Johnson, K.S.J. Role of the ubiquitin proteasome system in hematologic malignancies. Immunol. Rev. 2015, 263, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Shirazi, F.; Singh, R.K.; Kuiatse, I.; Wang, H.; Lee, H.C.; Berkova, Z.; Berger, A.; Hyer, M.; Chattopadhyay, N.; et al. Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood 2019, 133, 1572–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin–proteasome system as drug targets. Nat. Rev. Drug Discov. 2010, 10, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, D.; Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 2009, 458, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Chiba, T.; Tatsumi, K.; Iemura, S.; Tanida, I.; Okazaki, N.; Ueno, T.; Kominami, E.; Natsume, T.; Tanaka, K. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004, 23, 1977–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinform. 2016, 14, 140–146. [Google Scholar] [Green Version]
- Yoo, H.M.; Kang, S.H.; Kim, J.Y.; Lee, J.E.; Seong, M.W.; Lee, S.W.; Ka, S.H.; Sou, Y.-S.; Komatsu, M.; Tanaka, K.; et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 2014, 56, 261–274. [Google Scholar] [CrossRef]
- Gavin, J.M.; Hoar, K.; Xu, Q.; Ma, J.; Lin, Y.; Chen, J.; Chen, W.; Bruzzese, F.J.; Harrison, S.; Mallender, W.D.; et al. Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway. J. Biol. Chem. 2014, 289, 22648–22658. [Google Scholar] [CrossRef]
- da Silva, S.R.; Paiva, S.-L.; Bancerz, M.; Geletu, M.; Lewis, A.M.; Chen, J.; Cai, Y.; Lukkarila, J.L.; Li, H.; Gunning, P.T. A selective inhibitor of the UFM1-activating enzyme, UBA5. Bioorganic. Med. Chem. Lett. 2016, 26, 4542–4547. [Google Scholar] [CrossRef]
- Monks, A.; Scudiero, D.; Skehan, P.; Shoemaker, R.; Paull, K.; Vistica, D.; Hose, C.; Langley, J.; Cronise, P.; Vaigro-Wolff, A.; et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. JNCI J. Natl. Cancer Inst. 1991, 83, 757–766. [Google Scholar] [CrossRef]
- Seeliger, D.; Haas, J.; de Groot, B.L. Geometry-based sampling of conformational transitions in proteins. Structure 2007, 15, 1482–1492. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Shen, M.; Tian, S.; Xu, L.; Pan, P.; Guan, Y.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys. Chem. Chem. Phys. 2014, 16, 22035–22045. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S.M.; Sethi, G.; Lee, G.S.; Malek, S.N.A. Chalepin: Isolated from Ruta angustifolia L. Pers induces mitochondrial mediated apoptosis in lung carcinoma cells. BMC Complement. Altern. Med. 2016, 16, 389. [Google Scholar] [CrossRef] [PubMed]
- Sandjo, L.P.; Kuete, V.; Tchangna, R.S.; Efferth, T.; Ngadjui, B.T. Cytotoxic benzophenanthridine and furoquinoline alkaloids from Zanthoxylum buesgenii (Rutaceae). Chem. Cent. J. 2014, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Nganou, B.K.; Mbaveng, A.T.; Fobofou, S.A.T.; Fankam, A.G.; Bitchagno, G.T.M.; Simo Mpetga, J.D.; Wessjohann, L.A.; Kuete, V.; Efferth, T.; Tane, P. Furoquinolines and dihydrooxazole alkaloids with cytotoxic activity from the stem bark of Araliopsis soyauxii. Fitoterapia 2019, 133, 193–199. [Google Scholar] [CrossRef]
- Prakash Chaturvedula, V.S.; Schilling, J.K.; Miller, J.S.; Andriantsiferana, R.; Rasamison, V.E.; Kingston, D.G.I. New Cytotoxic Alkaloids from the wood of Vepris punctata from the Madagascar rainforest. J. Nat. Prod. 2003, 66, 532–534. [Google Scholar] [CrossRef]
- Chen, H.; Li, S.; Wang, S.; Li, W.; Bao, N.; Ai, W. The inhibitory effect of kokusaginine on the growth of human breast cancer cells and MDR-resistant cells is mediated by the inhibition of tubulin assembly. Bioorganic Med. Chem. Lett. 2018, 28, 2490–2492. [Google Scholar] [CrossRef]
- Cui, B.; Chai, H.; Dong, Y.; Horgen, F.D.; Hansen, B.; Madulid, D.A.; Soejarto, D.D.; Farnsworth, N.R.; Cordell, G.A.; Pezzuto, J.M.; et al. Quinoline alkaloids from Acronychia laurifolia. Phytochemistry 1999, 52, 95–98. [Google Scholar] [CrossRef]
- Best, S.; Liu, T.; Bruss, N.; Kittai, A.; Berger, A.; Danilov, A.V. Pharmacologic inhibition of the ubiquitin-activating enzyme induces ER stress and apoptosis in chronic lymphocytic leukemia and ibrutinib-resistant mantle cell lymphoma cells. Leuk. Lymphoma 2019, 1–5. [Google Scholar] [CrossRef]
- Cai, Y.; Pi, W.; Sivaprakasam, S.; Zhu, X.; Zhang, M.; Chen, J.; Makala, L.; Lu, C.; Wu, J.; Teng, Y.; et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for Ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015, 11, e1005643. [Google Scholar] [CrossRef] [PubMed]
- Hyer, M.L.; Milhollen, M.A.; Ciavarri, J.; Fleming, P.; Traore, T.; Sappal, D.; Huck, J.; Shi, J.; Gavin, J.; Brownell, J.; et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018, 24, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Bacik, J.-P.; Walker, J.R.; Ali, M.; Schimmer, A.D.; Dhe-Paganon, S. Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: Mechanistic insights into a minimalistic E1 enzyme. J. Biol. Chem. 2010, 285, 20273–20280. [Google Scholar] [CrossRef] [PubMed]



| Treatment | Cell Inhibition (%) | ||||||
|---|---|---|---|---|---|---|---|
| Cancer Cell Lines g | |||||||
| U251 | PC-3 | K562 | HCT-15 | MCF-7 | SKLU-1 | MRC5 | |
| EEa a | 29.5 ± 2.5 | 68.3 ± 1.2 | 96.8 ± 1.2 | 43.5 ± 2.7 | 62.7 ± 1.9 | 66.5 ± 4.5 | 18.3 ± 2.1 |
| CEa b | 27.4 ± 3.1 | 67.9 ± 1.5 | 97.2 ± 0.9 | 48.7 ± 2.1 | 45.7 ± 2.4 | 60.3 ± 3.1 | 21.3 ± 1.8 |
| EA-EEa3 c | 35.3 ± 2.7 | 61.4 ± 1.9 | 88.3 ± 1.8 | 44.5 ± 2.7 | 48.4 ± 3.3 | 31.5 ± 2.8 | 11.4 ± 1.2 |
| EA-EEa6 d | 30.2 ± 4.2 | 63.2 ± 2.2 | 91.7 ± 1.4 | 46.2 ± 1.2 | 36.5 ± 1.7 | 25.6 ± 3.2 | 19.9 ± 1.5 |
| 1 e | 31.9 ± 1.8 | 58.5 ± 2.0 | 86.7 ± 1.8 | 58.0 ± 3.2 | 31.9 ± 2.3 | 32.7 ± 1.7 | 5.3 ± 0.9 |
| 2 e | 43.4 ± 2.1 | 59.8 ± 1.7 | 93.4 ± 2.3 | 66.5 ± 2.5 | 52.8 ± 2.9 | 30.8 ± 2.2 | 4.3 ± 1.1 |
| doxorubicin f | 98.7 ± 0.9 | 82.1 ± 1.8 | 91.4 ± 1.9 | 93.2 ± 2.5 | 87.0 ± 3.1 | 90.8 ± 2.8 | 77.4 ± 3.2 |
| Complex | Evdw | Eelec | Gpolar | Gnonpolar | ΔGbinding |
|---|---|---|---|---|---|
| UBA5—1 | −145.3 ± 5.1 | −17.8 ± 3.8 | 75.6 ± 7.5 | −15.4 ± 0.5 | −102.9 ± 5.7 |
| UBA5—2 | −156.2 ± 7.9 | −16.3 ± 7.1 | 73.9 ± 17.2 | −15.9 ± 0.5 | −114.5 ± 4.1 |
| UBA5—TAK-243 | −186.7 ± 6.3 | −88.4 ± 12.6 | 222.7 ± 7.2 | −19.2 ± 0.6 | −71.6 ± 7.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Caballero, J.M.; Cuca-Suárez, L.E.; Coy-Barrera, E. Bio-Guided Fractionation of Ethanol Extract of Leaves of Esenbeckia alata Kunt (Rutaceae) Led to the Isolation of Two Cytotoxic Quinoline Alkaloids: Evidence of Selectivity Against Leukemia Cells. Biomolecules 2019, 9, 585. https://doi.org/10.3390/biom9100585
Álvarez-Caballero JM, Cuca-Suárez LE, Coy-Barrera E. Bio-Guided Fractionation of Ethanol Extract of Leaves of Esenbeckia alata Kunt (Rutaceae) Led to the Isolation of Two Cytotoxic Quinoline Alkaloids: Evidence of Selectivity Against Leukemia Cells. Biomolecules. 2019; 9(10):585. https://doi.org/10.3390/biom9100585
Chicago/Turabian StyleÁlvarez-Caballero, Juan Manuel, Luis Enrique Cuca-Suárez, and Ericsson Coy-Barrera. 2019. "Bio-Guided Fractionation of Ethanol Extract of Leaves of Esenbeckia alata Kunt (Rutaceae) Led to the Isolation of Two Cytotoxic Quinoline Alkaloids: Evidence of Selectivity Against Leukemia Cells" Biomolecules 9, no. 10: 585. https://doi.org/10.3390/biom9100585