Non-Canonical Amino Acids as Building Blocks for Peptidomimetics: Structure, Function, and Applications
Abstract
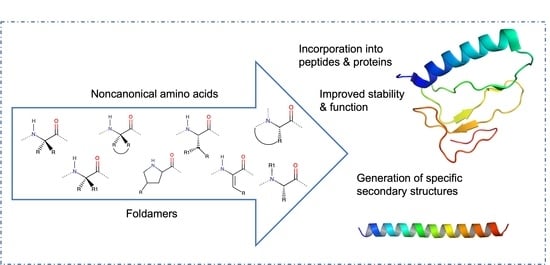
:1. Introduction
Amino Acids and Peptides: Structural Features and Properties
2. Peptidomimetics Design
2.1. Structural Properties of Non-Canonical Amino Acids
2.1.1. Symmetrical and Asymmetrical α,α-Dialkyl Glycines
2.1.2. Cα to Cα Cyclized Amino Acids—Acnc Residues
2.1.3. Proline Analogues
2.1.4. β-Substituted and Planar Amino Acids
2.1.5. α,β-Dehydroamino Acids
2.1.6. N-Cyclization and N-Alkylation
2.1.7. Other Side-Chain Modified Amino Acids
2.2. Backbone Modifications
3. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antosova, Z.; Mackova, M.; Kral, V.; Macek, T. Therapeutic Application of Peptides and Proteins: Parenteral Forever? Trends Biotechnol. 2009, 27, 628–635. [Google Scholar] [CrossRef]
- Kieber-Emmons, T.; Murali, R.; Greene, M.I. Therapeutic Peptides and Peptidomimetics. Curr. Opin. Biotechnol. 1997, 8, 435–441. [Google Scholar] [CrossRef]
- Lien, S.; Lowman, H.B. Therapeutic Peptides. Trends Biotechnol. 2003, 21, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B.-M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic Therapeutic Peptides: Science and Market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical Modifications Designed to Improve Peptide Stability: Incorporation of Non-Natural Amino Acids, Pseudo-Peptide Bonds, and Cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Lee, V.H.L.; Yamamoto, A. Penetration and Enzymatic Barriers to Peptide and Protein Absorption. Adv. Drug Deliv. Rev. 1989, 4, 171–207. [Google Scholar] [CrossRef]
- Bocci, V. Catabolism of Therapeutic Proteins and Peptides with Implications for Drug Delivery. Adv. Drug Deliv. Rev. 1989, 4, 149–169. [Google Scholar] [CrossRef]
- Lenci, E.; Trabocchi, A. Peptidomimetic Toolbox for Drug Discovery. Chem. Soc. Rev. 2020, 49, 3262–3277. [Google Scholar] [CrossRef]
- Avan, I.; Hall, C.D.; Katritzky, A.R. Peptidomimetics via Modifications of Amino Acids and Peptide Bonds. Chem. Soc. Rev. 2014, 43, 3575. [Google Scholar] [CrossRef]
- Grauer, A.; König, B. Peptidomimetics—A Versatile Route to Biologically Active Compounds. Eur. J. Org. Chem. 2009, 2009, 5099–5111. [Google Scholar] [CrossRef]
- Gentilucci, L.; Tolomelli, A.; Squassabia, F. Peptides and Peptidomimetics in Medicine, Surgery and Biotechnology. Curr. Med. Chem. 2006, 13, 2449–2466. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic Peptides: Current Applications and Future Directions. Sig. Transduct. Target Ther. 2022, 7, 1–27. [Google Scholar] [CrossRef]
- Vagner, J.; Qu, H.; Hruby, V.J. Peptidomimetics, a Synthetic Tool of Drug Discovery. Curr. Opin. Chem. Biol. 2008, 12, 292–296. [Google Scholar] [CrossRef] [Green Version]
- Crisma, M.; De Zotti, M.; Formaggio, F.; Peggion, C.; Moretto, A.; Toniolo, C. Handedness Preference and Switching of Peptide Helices. Part II: Helices Based on Noncoded α -Amino Acids: PEPTIDE HELIX HANDEDNESS (II). J. Pept. Sci. 2015, 21, 148–177. [Google Scholar] [CrossRef]
- Connor, R.E.; Tirrell, D.A. Non-Canonical Amino Acids in Protein Polymer Design. Polym. Rev. 2007, 47, 9–28. [Google Scholar] [CrossRef]
- Do, T.; Link, A.J. Protein Engineering in Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs). Biochemistry 2023, 62, 201–209. [Google Scholar] [CrossRef]
- Qvit, N.; Rubin, S.J.S.; Urban, T.J.; Mochly-Rosen, D.; Gross, E.R. Peptidomimetic Therapeutics: Scientific Approaches and Opportunities. Drug Discov. Today 2017, 22, 454–462. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Willi, J.A.; Cho, N.; Kim, I.; Jewett, M.C.; Lee, J. Cell-Free Biosynthesis of Peptidomimetics. Biotechnol. Bioproc. E 2023, 1–17. [Google Scholar] [CrossRef]
- Du, Y.; Li, L.; Zheng, Y.; Liu, J.; Gong, J.; Qiu, Z.; Li, Y.; Qiao, J.; Huo, Y.-X. Incorporation of Non-Canonical Amino Acids into Antimicrobial Peptides: Advances, Challenges, and Perspectives. Appl. Environ. Microbiol. 2022, 88, e01617-22. [Google Scholar] [CrossRef]
- Singh, S.; Singh, H.; Tuknait, A.; Chaudhary, K.; Singh, B.; Kumaran, S.; Raghava, G.P.S. PEPstrMOD: Structure Prediction of Peptides Containing Natural, Non-Natural and Modified Residues. Biol. Direct. 2015, 10, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhadi, T.; Hashemian, S.M. Computer-Aided Design of Amino Acid-Based Therapeutics: A Review. DDDT 2018, 12, 1239–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reese, H.R.; Shanahan, C.C.; Proulx, C.; Menegatti, S. Peptide Science: A “Rule Model” for New Generations of Peptidomimetics. Acta Biomater. 2020, 102, 35–74. [Google Scholar] [CrossRef] [PubMed]
- D’Annessa, I.; Di Leva, F.S.; La Teana, A.; Novellino, E.; Limongelli, V.; Di Marino, D. Bioinformatics and Biosimulations as Toolbox for Peptides and Peptidomimetics Design: Where Are We? Front. Mol. Biosci. 2020, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.L.; Miller, S.L. Reasons for the Occurrence of the Twenty Coded Protein Amino Acids. J. Mol. Evol. 1981, 17, 273–284. [Google Scholar] [CrossRef]
- Blumenkrantz, N.; Ullman, S.; Asboe-Hansen, G. Parallel Studies on Collagen Hydroxyproline and Hydroxylysine in Human Skin Biopsies. Acta Derm. Venereol. 1976, 56, 93–98. [Google Scholar]
- Gorres, K.L.; Raines, R.T. Prolyl 4-Hydroxylase. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 106–124. [Google Scholar] [CrossRef]
- Szpak, P. Fish Bone Chemistry and Ultrastructure: Implications for Taphonomy and Stable Isotope Analysis. J. Archaeol. Sci. 2011, 38, 3358–3372. [Google Scholar] [CrossRef]
- Zhu, X.; Tang, G.; Galili, G. The Catabolic Function of the α-Aminoadipic Acid Pathway in Plants Is Associated with Unidirectional Activity of Lysine-Oxoglutarate Reductase, but Not Saccharopine Dehydrogenase. Biochem. J. 2000, 351, 215–220. [Google Scholar] [CrossRef]
- Peisach, J.; Blumberg, W.E. A Mechanism for the Action of Penicillamine in the Treatment of Wilson’s Disease. Mol. Pharmacol. 1969, 5, 200–209. [Google Scholar]
- Sugino, T.; Shirai, T.; Kajimoto, Y.; Kajimoto, O. L-Ornithine Supplementation Attenuates Physical Fatigue in Healthy Volunteers by Modulating Lipid and Amino Acid Metabolism. Nutr. Res. 2008, 28, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Fearon, W.R. The Carbamido Diacetyl Reaction: A Test for Citrulline. Biochem. J. 1939, 33, 902–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rother, M.; Krzycki, J.A. Selenocysteine, Pyrrolysine, and the Unique Energy Metabolism of Methanogenic Archaea. Archaea 2010, 2010, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gladyshev, V.N. High Content of Proteins Containing 21st and 22nd Amino Acids, Selenocysteine and Pyrrolysine, in a Symbiotic Deltaproteobacterium of Gutless Worm Olavius Algarvensis. Nucleic Acids Res. 2007, 35, 4952–4963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaston, M.A.; Jiang, R.; Krzycki, J.A. Functional Context, Biosynthesis, and Genetic Encoding of Pyrrolysine. Curr. Opin. Microbiol. 2011, 14, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Johansson, L.; Gafvelin, G.; Arnér, E.S.J. Selenocysteine in Proteins—Properties and Biotechnological Use. Biochim. Et Biophys. Acta (BBA)—Gen. Subj. 2005, 1726, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranov, P.V.; Gesteland, R.F.; Atkins, J.F. Recoding: Translational Bifurcations in Gene Expression. Gene 2002, 286, 187–201. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Korniy, N.; Klimova, M.; Karki, P.; Peng, B.-Z.; Senyushkina, T.; Belardinelli, R.; Maracci, C.; Wohlgemuth, I.; Samatova, E.; et al. Translational Recoding: Canonical Translation Mechanisms Reinterpreted. Nucleic. Acids Res. 2020, 48, 1056–1067. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.; Cox, M. Lehninger: Principles of Biochemistry, 7th ed.Palgrave W. H. Freeman: New York, NY, USA, 2014. [Google Scholar]
- Welker, M.; Von Döhren, H. Cyanobacterial Peptides—Nature’s Own Combinatorial Biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563. [Google Scholar] [CrossRef] [Green Version]
- Chlipala, G.E.; Mo, S.; Orjala, J. Chemodiversity in Freshwater and Terrestrial Cyanobacteria—A Source for Drug Discovery. Current. Drug Targets 2011, 12, 1654–1673. [Google Scholar] [CrossRef]
- Martinovich, V.P.; Baradzina, K.U. Peptide Hormones in Medicine: A 100-Year History. Russ. J. Bioorg. Chem. 2022, 48, 221–232. [Google Scholar] [CrossRef]
- Lee, H.-J.; Macbeth, A.H.; Pagani, J.H.; Scott Young, W. Oxytocin: The Great Facilitator of Life. Prog. Neurobiol. 2009, 88, 127–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urayama, A.; Yamada, S.; Kimura, R.; Zhang, J.; Watanabe, Y. Neuroprotective Effect and Brain Receptor Binding of Taltirelin, a Novel Thyrotropin-Releasing Hormone (TRH) Analogue, in Transient Forebrain Ischemia of C57BL/6J Mice. Life Sci. 2002, 72, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, I.; Separovic, F.; Auger, M.; Gagné, S.M. A Multidimensional 1H NMR Investigation of the Conformation of Methionine-Enkephalin in Fast-Tumbling Bicelles. Biophys. J. 2004, 86, 1587–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovmöller, S.; Zhou, T.; Ohlson, T. Conformations of Amino Acids in Proteins. Acta Cryst. D 2002, 58, 768–776. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of Polypeptide Chain Configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, P.; Epand, R.F.; Heidenreich, N.; Bürck, J.; Ulrich, A.S.; Epand, R.M. Membrane-Active Peptides and the Clustering of Anionic Lipids. Biophys. J. 2012, 103, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendel, D.; Ellman, J.; Schultz, P.G. Protein Biosynthesis with Conformationally Restricted Amino Acids. J. Am. Chem. Soc. 1993, 115, 4359–4360. [Google Scholar] [CrossRef]
- Giannis, A.; Rübsam, F. Peptidomimetics in Drug Design. In Advances in Drug Research; Testa, B., Meyer, U.A., Eds.; Academic Press: Cambridge, MA, USA, 1997; Volume 29, pp. 1–78. [Google Scholar]
- Whitmore, L.; Wallace, B.A. The Peptaibol Database: A Database for Sequences and Structures of Naturally Occurring Peptaibols. Nucleic. Acids Res. 2004, 32, D593–D594. [Google Scholar] [CrossRef] [Green Version]
- Siodłak, D. α,β-Dehydroamino Acids in Naturally Occurring Peptides. Amino. Acids 2015, 47, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Domalaon, R.; GZhanel, G.; Schweizer, F. Short Antimicrobial Peptides and Peptide Scaffolds as Promising Antibacterial Agents. Curr. Top. Med. Chem. 2016, 16, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ben Haj Salah, K.; Djibo, M.; Inguimbert, N. Peptaibols as a Model for the Insertions of Chemical Modifications. Arch. Biochem. Biophys. 2018, 658, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Chugh, J.K.; Wallace, B.A. Peptaibols: Models for Ion Channels. Biochem. Soc. Trans. 2001, 29, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Brückner, H. Peptaibiomics: Towards a Myriad of Bioactive Peptides Containing Cα-Dialkylamino Acids? Chem. Biodivers. 2008, 5, 1817–1843. [Google Scholar] [CrossRef] [PubMed]
- Duclohier, H. Peptaibiotics and Peptaibols: An Alternative to Classical Antibiotics? Chem. Biodivers. 2007, 4, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Karle, I.L.; Balaram, P. Structural Characteristics of.Alpha.-Helical Peptide Molecules Containing Aib Residues. Biochemistry 1990, 29, 6747–6756. [Google Scholar] [CrossRef]
- Toniolo, C.; Crisma, M.; Formaggio, F.; Valle, C.; Cavicchioni, G.; Precigoux, G.; Aubry, A.; Kamphuis, J. Structures of Peptides from A-amino Acids Methylated at the A-carbon. Biopolymers 1993, 33, 1061–1072. [Google Scholar] [CrossRef]
- Castro, T.G.; Micaêlo, N.M. Modeling of Peptaibol Analogues Incorporating Nonpolar α,α-Dialkyl Glycines Shows Improved α-Helical Preorganization and Spontaneous Membrane Permeation. J. Phys. Chem. B 2014, 118, 649–658. [Google Scholar] [CrossRef]
- Castro, T.G.; Micaêlo, N.M. Conformational and Thermodynamic Properties of Non-Canonical α,α-Dialkyl Glycines in the Peptaibol Alamethicin: Molecular Dynamics Studies. J. Phys. Chem. B 2014, 118, 9861–9870. [Google Scholar] [CrossRef] [PubMed]
- Castro, T.G.; Micaêlo, N.M.; Melle-Franco, M. Modeling the Secondary Structures of the Peptaibols Antiamoebin I and Zervamicin II Modified with D-Amino Acids and Proline Analogues. J. Mol. Model. 2017, 23, 313. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.I.B.; Carvalho, C.M.; Fernandes, R.D.V.; Pereira-Lima, S.M.M.A.; Castanheira, E.M.S.; Costa, S.P.G. Peptaibolin Analogues by Incorporation of α,α-Dialkylglycines: Synthesis and Study of Their Membrane Permeating Ability. Tetrahedron 2016, 72, 1024–1030. [Google Scholar] [CrossRef]
- Nagaraj, R.; Balaram, P. A Stereochemically-Constrained Enkephalin Analog: α-Aminoisobutyryl 2 Methionine 5 Enkephalinamide. FEBS Lett. 1978, 96, 273–276. [Google Scholar] [CrossRef] [Green Version]
- Nagaraj, R.; Sudha, T.S.; Shivaji, S.; Balaram, P. Enkephalin Analogs. Introduction of Stereochemical Constraints, Metal Binding Sites and Fluorescent Groups. FEBS Lett. 1979, 106, 271–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vavrek, R.J.; Stewart, J.M. Bradykinin Analogs Containing α-Aminoisobutyric Acid (Aib). Peptides 1980, 1, 231–235. [Google Scholar] [CrossRef]
- Cordopatis, P.; Theodoropoulos, D. Synthesis of [1-Aib]-Angiotensin II, an Analogue with Higher Potency than [1-Asn,5-Val]-Angiotensin II. Experientia 1983, 39, 106–108. [Google Scholar] [CrossRef]
- Stewart, J.M. Bradykinin Antagonists: Discovery and Development. Peptides 2004, 25, 527–532. [Google Scholar] [CrossRef]
- Doi, M.; Asano, A.; Komura, E.; Ueda, Y. The Structure of an Endomorphin Analogue Incorporating 1-Aminocyclohexane-1-Carboxlylic Acid for Proline Is Similar to the b-Turn of Leu-Enkephalin. Biochem. Biophys. Res. Commun. 2002, 5, 138–142. [Google Scholar] [CrossRef]
- Toniolo, C.; Crisma, M.; Valle, G.; Bonora, G.M.; Polinelli, S.; Becker, E.L.; Freer, R.J.; Sudhanand; Rao, R.B.; Balaram, P. Conformationally Restricted Formyl Methionyl Tripeptide Chemoattractants: A Three-Dimensional Structure-Activity Study of Analogs Incorporating a C Alpha, Alpha-Dialkylated Glycine at Position 2. Pept. Res. 1989, 2, 275–281. [Google Scholar]
- Roos, E.C.; Lopez, M.C.; Brook, M.A.; Hiemstra, H.; Speckamp, W.N.; Kaptein, B.; Kamphuis, J.; Schoemaker, H.E. Synthesis of.Alpha.-Substituted.Alpha.-Amino Acids via Cationic Intermediates. J. Org. Chem. 1993, 58, 3259–3268. [Google Scholar] [CrossRef]
- De Zotti, M.; Sella, L.; Bolzonello, A.; Gabbatore, L.; Peggion, C.; Bortolotto, A.; Elmaghraby, I.; Tundo, S.; Favaron, F. Targeted Amino Acid Substitutions in a Trichoderma Peptaibol Confer Activity against Fungal Plant Pathogens and Protect Host Tissues from Botrytis Cinerea Infection. Int. J. Mol. Sci. 2020, 21, 7521. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Balashova, T.A.; Efremov, R.G.; Yakimenko, Z.A.; Ovchinnikova, T.V.; Raap, J.; Arseniev, A.S. Spatial Structure of Zervamicin IIB Bound to DPC Micelles: Implications for Voltage-Gating. Biophys. J. 2002, 82, 762–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snook, C.; Woolley, G.; Oliva, G.; Pattabhi, V.; Wood, S.; Blundell, T.; Wallace, B. The Structure and Function of Antiamoebin I, a Proline-Rich Membrane-Active Polypeptide. Structure 1998, 6, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, E.; Blasio, B.D.; Iacovino, R.; Menchise, V.; Saviano, M.; Pedone, C.; Bonora, G.M.; Ettorre, A.; Graci, L.; Formaggio, F.; et al. Conformational Restriction through C α i ←→ C α i Cyclization: 1-Aminocycloheptane-1-Carboxylic Acid (Ac 7 c). J. Chem. Soc. Perkin Trans. 2 1997, 0, 2023–2032. [Google Scholar] [CrossRef]
- Toniolo, C. Conformationally Restricted Peptides through Short-Range Cyclizations. Int. J. Pept. Protein Res. 1990, 35, 287–300. [Google Scholar] [CrossRef]
- Aschi, M.; Lucente, G.; Mazza, F.; Mollica, A.; Morera, E.; Nalli, M.; Paradisi, M.P. Peptide Backbone Folding Induced by the Cα -Tetrasubstituted Cyclic -Amino Acids 4-Amino-1,2-Dithiolane-4-Carboxylic Acid (Adt) and 1-Aminocyclopentane-1-Carboxylic Acid (Ac5c). A Joint Computational and Experimental Study. Org. Biomol. Chem. 2003, 1, 1980–1988. [Google Scholar] [CrossRef]
- Bardi, R.; Piazzesi, A.M.; Toniolo, C.; Sukumar, M.; Balaram, P. Stereochemistry of Peptides Containing 1-Aminocyclopentanecarboxylic Acid (Acc5): Solution and Solid-State Conformations of Boc-Acc5-Acc5-NHMe. Biopolymers 1986, 25, 1635–1644. [Google Scholar] [CrossRef]
- Barone, V.; Fraternali, F.; Cristinziano, P.L.; Lelj, F.; Rosa, A. Conformational Behavior of α,α-Dialkylated Peptides: Ab Initio and Empirical Results for Cyclopropylglycine. Biopolymers 1988, 27, 1673–1685. [Google Scholar] [CrossRef]
- Demizu, Y.; Doi, M.; Kurihara, M.; Okuda, H.; Nagano, M.; Suemune, H.; Tanaka, M. Conformational Studies on Peptides Containing α,α-Disubstituted α-Amino Acids: Chiral Cyclic α,α-Disubstituted α-Amino Acid as an α-Helical Inducer. Org. Biomol. Chem. 2011, 9, 3303–3312. [Google Scholar] [CrossRef]
- Di Blasio, B.; Lombardi, A.; Nastri, F.; Saviano, M.; Pedone, C.; Yamada, T.; Nakao, M.; Kuwata, S.; Pavone, V. Conformation of Diastereomeric Peptide Sequences: Structural Analysis of Z-D-Val-Ac6c-Gly-L-Phe-OMe. Biopolymers 1992, 32, 1155–1161. [Google Scholar] [CrossRef]
- Gatos, M.; Formaggio, F.; Crisma, M.; Toniolo, C.; Bonora, G.M.; Benedetti, Z.; Di Blasio, B.; Iacovino, R.; Santini, A.; Saviano, M.; et al. Conformational Characterization of the 1-Aminocyclobutane-1-Carboxylic Acid Residue in Model Peptides. J. Pept. Sci. 1997, 3, 110–122. [Google Scholar] [CrossRef]
- Ballano, G.; Zanuy, D.; Jiménez, A.I.; Cativiela, C.; Nussinov, R.; Alemán, C. Structural Analysis of a β-Helical Protein Motif Stabilized by Targeted Replacements with Conformationally Constrained Amino Acids. J. Phys. Chem. B 2008, 112, 13101–13115. [Google Scholar] [CrossRef] [Green Version]
- Gatos, M.; Formaggio, F.; Crisma, M.; Valle, G.; Toniolo, C.; Bonora, G.M.; Saviano, M.; Iacovino, R.; Menchise, V.; Galdiero, S.; et al. Conformational Characterization of Peptides Rich in the Cycloaliphatic Cα,α-Disubstituted Glycine 1-Aminocyclononane-1-Carboxylic Acid. J. Pept. Sci. 1997, 3, 367–382. [Google Scholar] [CrossRef]
- Moretto, A.; Formaggio, F.; Crisma, M.; Toniolo, C.; Saviano, M.; Benedetti, E.; Iacovino, R.; Vitale, R.M. Ac10c: A Medium-Ring, Cycloaliphatic Cα,α-Disubstituted Glycine. Incorporation into Model Peptides and Preferred Conformation. J. Pept. Res. 2001, 57, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Saviano, M.; Iacovino, R.; Benedetti, E.; Moretto, V.; Banzato, A.; Formaggio, F.; Crisma, M.; Toniolo, C. Preferred Conformation of Peptides Based on Cycloaliphatic Cα,α-Disubstituted Glycines: 1-Amino-Cycloundecane-1-Carboxylic Acid (Ac11c). J. Pept. Sci. 2000, 6, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Saviano, M.; Iacovino, R.; Menchise, V.; Benedetti, E.; Bonora, G.M.; Gatos, M.; Graci, L.; Formaggio, F.; Crisma, M.; Toniolo, C. Conformational Restriction through C ↔ C Cyclization: Ac12c, the Largest Cycloaliphatic Cα,α- Disubstituted Glycine Known. Biopolymers 2000, 53, 200–212. [Google Scholar] [CrossRef]
- Paglialunga Paradisi, M.; Torrini, I.; Pagani Zecchini, G.; Lucente, G.; Gavuzzo, E.; Mazza, F.; Pochetti, G. γ-Turn Conformation Induced by α,α-Disubstituted Amino Acids with a Cyclic Six-Membered Side Chain. Tetrahedron 1995, 51, 2379–2386. [Google Scholar] [CrossRef]
- Rodríguez-Ropero, F.; Zanuy, D.; Casanovas, J.; Nussinov, R.; Alemán, C. Application of 1-Aminocyclohexane Carboxylic Acid to Protein Nanostructure Computer Design. J. Chem. Inf. Model. 2008, 48, 333–343. [Google Scholar] [CrossRef]
- Alemán, C. Conformational Properties of α-Amino Acids Disubstituted at the α-Carbon. J. Phys. Chem. B 1997, 101, 5046–5050. [Google Scholar] [CrossRef]
- Benedetti, E.; Di Blasio, B.; Pavone, V.; Pedone, C.; Santini, A.; Barone, V.; Fraternali, F.; Lelj, F.; Bavoso, A.; Crisma, M.; et al. Structural Versatility of Peptides Containing Cα,α-Dialkylated Glycines. An X-Ray Diffraction Study of Six 1-Aminocyclopropane-1-Carboxylic Acid Rich Peptides. Int. J. Biol. Macromol. 1989, 11, 353–360. [Google Scholar] [CrossRef]
- Jiménez, A.I.; Vaquero, V.; Cabezas, C.; López, J.C.; Cativiela, C.; Alonso, J.L. The Singular Gas-Phase Structure of 1-Aminocyclopropanecarboxylic Acid (Ac3c). J. Am. Chem. Soc. 2011, 133, 10621–10628. [Google Scholar] [CrossRef]
- Gomez-Catalan, J.; Aleman, C.; Perez, J.J. Conformational Profile of 1-Aminocyclopropanecarboxylic Acid. Theor. Chem. Acc. 2000, 103, 380–389. [Google Scholar] [CrossRef]
- Headley, A.D.; Ganesan, R.; Nam, J. The Effect of the Cyclopropyl Group on the Conformation of Chemotactic Formyl Tripeptides. Bioorganic Chem. 2003, 31, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Zanuy, D.; Ballano, G.; Jiménez, A.I.; Casanovas, J.; Haspel, N.; Cativiela, C.; Curcó, D.; Nussinov, R.; Alemán, C. Protein Segments with Conformationally Restricted Amino Acids Can Control Supramolecular Organization at the Nanoscale. J. Chem. Inf. Model. 2009, 49, 1623–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atta-ur-Rahman, M.; Choudhary, I. (Eds.) Frontiers in Drug Design and Discovery: Volume 10; Frontiers in Drug Design and Discovery; Bentham Science Publishers: Sharjah, United Arab Emirates, 2021; Volume 1, ISBN 9789811421563. [Google Scholar]
- Baures, P.W.; Ojala, W.H.; Gleason, W.B.; Johnson, R.L. Conformational Analysis of Homochiral and Heterochiral Diprolines as β-Turn-Forming Peptidomimetics: Unsubstituted and Substituted Models. J. Pept. Res. 1997, 50, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Caumes, C.; Delsuc, N.; Azza, R.B.; Correia, I.; Chemla, F.; Ferreira, F.; Carlier, L.; Luna, A.P.; Moumné, R.; Lequin, O.; et al. Homooligomers of Substituted Prolines and β-Prolines: Syntheses and Secondary Structure Investigation. New J. Chem. 2013, 37, 1312–1319. [Google Scholar] [CrossRef]
- Bach, T.M.H.; Takagi, H. Properties, Metabolisms, and Applications of l-Proline Analogues. Appl. Microbiol. Biotechnol. 2013, 97, 6623–6634. [Google Scholar] [CrossRef]
- Tsogoeva, S.B.; Jagtap, S.B.; Ardemasova, Z.A. 4-Trans-Amino-Proline Based Di- and Tetrapeptides as Organic Catalysts for Asymmetric C–C Bond Formation Reactions. Tetrahedron Asymmetry 2006, 17, 989–992. [Google Scholar] [CrossRef]
- Torino, D.; Mollica, A.; Pinnen, F.; Feliciani, F.; Spisani, S.; Lucente, G. Novel Chemotactic For-Met-Leu-Phe-OMe (FMLF-OMe) Analogues Based on Met Residue Replacement by 4-Amino-Proline Scaffold: Synthesis and Bioactivity. Bioorganic Med. Chem. 2009, 17, 251–259. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen Structure and Stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [Green Version]
- Bilsky, E.J.; Qian, X.; Hruby, V.J.; Porreca, F. Antinociceptive Activity of [β-Methyl-2′,6′-Dimethyltyrosine1]-Substituted Cyclic [d-Pen2,d-Pen5]Enkephalin and [d-Ala2,Asp4]Deltorphin Analogs. J. Pharmacol. Exp. Ther. 2000, 293, 151–158. [Google Scholar] [PubMed]
- Haskell-Luevano, C.; Toth, K.; Boteju, L.; Job, C.; Castrucci, A.M.D.L.; Hadley, M.E.; Hruby, V.J. β-Methylation of the Phe7 and Trp9 Melanotropin Side Chain Pharmacophores Affects Ligand−Receptor Interactions and Prolonged Biological Activity. J. Med. Chem. 1997, 40, 2740–2749. [Google Scholar] [CrossRef]
- Jiao, D.; Russell, K.C.; Hruby, V.J. Locally Constrained Tyrosine Analogues with Restricted Side Chain Dynamics. Tetrahedron 1993, 49, 3511–3520. [Google Scholar] [CrossRef]
- Kover, K.E.; Jiao, D.; Fang, S.; Hruby, V.J. Conformational Properties of the Unnatural Amino Acid.Beta.-Methylphenylalanine in a Linear Octapeptide System; Correlations of 13C-NMR Chemical Shifts with the Side-Chain Stereochemistry of These Amino Acid Residues. J. Org. Chem. 1994, 59, 991–998. [Google Scholar] [CrossRef]
- Mosberg, H.I.; Omnaas, J.R.; Lomize, A.; Heyl, D.L.; Nordan, I.; Mousigian, C.; Davis, P.; Porreca, F. Development of a Model for the Delta Opioid Receptor Pharmacophore. 2. Conformationally Restricted Phe3 Replacements in the Cyclic Delta Receptor Selective Tetrapeptide Tyr-c[D-Cys-Phe-D-Pen]OH (JOM-13). J. Med. Chem. 1994, 37, 4384–4391. [Google Scholar] [CrossRef] [PubMed]
- Hruby, V.J. Peptide Science: Exploring the Use of Chemical Principles and Interdisciplinary Collaboration for Understanding Life Processes. J. Med. Chem. 2003, 46, 4215–4231. [Google Scholar] [CrossRef] [PubMed]
- Hruby, V.J.; Cai, M. Design of Peptide and Peptidomimetic Ligands with Novel Pharmacological Activity Profiles. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 557–580. [Google Scholar] [CrossRef] [Green Version]
- Hruby, V.J. Conformational and Topographical Considerations in the Design of Biologically Active Peptides. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/bip.360330709 (accessed on 28 June 2019).
- Singh, T.P.; Kaur, P. Conformation and Design of Peptides with α,β-Dehydro-Amino Acid Residues. Prog. Biophys. Mol. Biol. 1996, 66, 141–165. [Google Scholar] [CrossRef]
- Singh, T.P.; Narula, P.; Patel, H.C. α,β-Dehydro Residues in the Design of Peptide and Protein Structures. Acta Crystallogr. B 1990, 46 Pt 4, 539–545. [Google Scholar] [CrossRef]
- Busetti, V.; Crisma, M.; Toniolo, C.; Salvadori, S.; Balboni, G. α,β-Dehydro-Amino Acid Residues in the Design of Peptide Structures. Molecular and Crystal Structures of Two Folded Dehydro Peptides. Int. J. Biol. Macromol. 1992, 14, 23–28. [Google Scholar] [CrossRef]
- Ciszak, E.; Pietrzyñski, G.; Rzeszotarska, B. Conformational Investigation of α, Β-dehydropeptides. Int. J. Pept. Protein Res. 1992, 39, 218–222. [Google Scholar] [CrossRef] [PubMed]
- English, M.L.; Stammer, C.H. The Enzyme Stability of Dehydropeptides. Biochem. Biophys. Res. Commun. 1978, 83, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Pieroni, O.; Fissi, A.; Jain, R.M.; Chauhan, V.S. Solution Structure of Dehydropeptides: A CD Investigation. Biopolymers 1996, 38, 97–108. [Google Scholar] [CrossRef]
- Scaloni, A.; Barra, D.; Bossa, F. Sequence Analysis of Dehydroamino Acid Containing Peptides. Anal. Biochem. 1994, 218, 226–228. [Google Scholar] [CrossRef]
- Oliveira, C.B.P.; Pereira, R.B.; Pereira, D.M.; Hilliou, L.; Castro, T.G.; Martins, J.A.; Jervis, P.J.; Ferreira, P.M.T. Aryl-Capped Lysine-Dehydroamino Acid Dipeptide Supergelators as Potential Drug Release Systems. IJMS 2022, 23, 11811. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.B.P.; Gomes, V.; Ferreira, P.M.T.; Martins, J.A.; Jervis, P.J. Peptide-Based Supramolecular Hydrogels as Drug Delivery Agents: Recent Advances. Gels 2022, 8, 706. [Google Scholar] [CrossRef]
- Joaquin, D.; Lee, M.A.; Kastner, D.W.; Singh, J.; Morrill, S.T.; Damstedt, G.; Castle, S.L. Impact of Dehydroamino Acids on the Structure and Stability of Incipient 310-Helical Peptides. J. Org. Chem. 2020, 85, 1601–1613. [Google Scholar] [CrossRef]
- Desai, P.V.; Coutinho, E.C. Effect of Stereochemistry (Z and E) and Position of α,β-Dehydrophenylalanine (ΔPhe) on β-Turn Stability. J. Mol. Model. 2000, 6, 595–599. [Google Scholar] [CrossRef]
- Gross, E.; Morell, J.L. Presence of Dehydroalanine in the Antibiotic Nisin and Its Relation to Activity. J. Am. Chem. Soc. 1967, 89, 2791–2792. [Google Scholar] [CrossRef]
- Nandel, F.S.; Sahrawat, T.R. Conformational Study of Poly-ΔAbu Peptides and Construction of Amphipathic Nanostructure. Pept. Sci. 2009, 92, 44–51. [Google Scholar] [CrossRef]
- Nandel, F.S.; Malik, N.; Singh, B.; Jain, D.V.S. Conformational Structure of Peptides Containing Dehydroalanine: Formation of β-Bend Ribbon Structure. Int. J. Quantum Chem. 1999, 72, 15–23. [Google Scholar] [CrossRef]
- Dey, S.; Mitra, S.N.; Singh, T.P. Design of Peptides Using α,β-Dehydro-Residues: Synthesis, Crystal Structure and Molecular Conformation of Boc-L-Val-Delta Phe-Delta Phe-L-Val-OCH3. Biopolymers 1996, 39, 849–857. [Google Scholar] [CrossRef]
- Seebach, D.; Beck, A.K.; Bierbaum, D.J. The World Ofβ- Andγ-Peptides Comprised of Homologated Proteinogenic Amino Acids and Other Components. Chem. Biodivers. 2004, 1, 1111–1239. [Google Scholar] [CrossRef]
- Siodłak, D.; Grondys, J.; Lis, T.; Bujak, M.; Broda, M.A.; Rzeszotarska, B. The Conformational Properties of Dehydrobutyrine and Dehydrovaline: Theoretical and Solid-State Conformational Studies. J. Pept. Sci. 2010, 16, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Buczek, A.; Siodłak, D.; Bujak, M.; Broda, M.A. Effects of Side-Chain Orientation on the Backbone Conformation of the Dehydrophenylalanine Residue. Theoretical and X-Ray Study. J. Phys. Chem. B 2011, 115, 4295–4306. [Google Scholar] [CrossRef]
- Yadav, S.; Bharti, S.; Srivastava, P.; Mathur, P. PepEngine: A Manually Curated Structural Database of Peptides Containing α, β- Dehydrophenylalanine (ΔPhe) and α-Amino Isobutyric Acid (Aib). Int. J. Pept. Res. Ther. 2022, 28, 57. [Google Scholar] [CrossRef]
- Rajashankar, K.R.; Ramakumar, S.; Chauhan, V.S. Design of a Helical Motif Using.Alpha.,.Beta.-Dehydrophenylalanine Residues: Crystal Structure of Boc-Val-.DELTA.Phe-Phe-Ala-Phe-.DELTA.Phe-Val-.DELTA.PHe-Gly-OCH3, a 310-Helical Nonapeptide. J. Am. Chem. Soc. 1992, 114, 9225–9226. [Google Scholar] [CrossRef]
- Mathur, P.; Ramakumar, S.; Chauhan, V.S. Peptide Design Using α,β-Dehydro Amino Acids: From Beta-Turns to Helical Hairpins. Biopolymers 2004, 76, 150–161. [Google Scholar] [CrossRef]
- Vilaça, H.; Pereira, G.; Castro, T.G.; Hermenegildo, B.F.; Shi, J.; Faria, T.Q.; Micaêlo, N.; Brito, R.M.M.; Xu, B.; Castanheira, E.M.S.; et al. New Self-Assembled Supramolecular Hydrogels Based on Dehydropeptides. J. Mater. Chem. B 2015, 3, 6355–6367. [Google Scholar] [CrossRef]
- Vilaça, H.; Castro, T.; Costa, F.M.G.; Melle-Franco, M.; Hilliou, L.; Hamley, I.W.; Castanheira, E.M.S.; Martins, J.A.; Ferreira, P.M.T. Self-Assembled RGD Dehydropeptide Hydrogels for Drug Delivery Applications. J. Mater. Chem. B 2017, 5, 8607–8617. [Google Scholar] [CrossRef] [Green Version]
- Molina-Guijarro, J.M.; García, C.; Macías, Á.; García-Fernández, L.F.; Moreno, C.; Reyes, F.; Martínez-Leal, J.F.; Fernández, R.; Martínez, V.; Valenzuela, C.; et al. Elisidepsin Interacts Directly with Glycosylceramides in the Plasma Membrane of Tumor Cells to Induce Necrotic Cell Death. PLoS ONE 2015, 10, e0140782. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Hamann, M.T. Chemistry and Biology of Kahalalides. Chem. Rev. 2011, 111, 3208–3235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyslop, J.F.; Lovelock, S.L.; Watson, A.J.B.; Sutton, P.W.; Roiban, G.-D. N-Alkyl-α-Amino Acids in Nature and Their Biocatalytic Preparation. J. Biotechnol. 2019, 293, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, J.A.E.; Kulkarni, K.; Izore, T.; Fulcher, A.J.; Peleg, A.Y.; Aguilar, M.-I.; Cryle, M.J.; Del Borgo, M.P. Staphylococcus Aureus Entanglement in Self-Assembling β-Peptide Nanofibres Decorated with Vancomycin. Nanoscale Adv. 2021, 3, 2607–2616. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhou, H.; Olademehin, O.P.; Kim, S.J.; Tao, P. Insights into Key Interactions between Vancomycin and Bacterial Cell Wall Structures. ACS Omega 2018, 3, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Kerbs, A.; Mindt, M.; Schwardmann, L.; Wendisch, V.F. Sustainable Production of N-Methylphenylalanine by Reductive Methylamination of Phenylpyruvate Using Engineered Corynebacterium Glutamicum. Microorganisms 2021, 9, 824. [Google Scholar] [CrossRef]
- Chatterjee, J.; Rechenmacher, F.; Kessler, H. N-Methylation of Peptides and Proteins: An Important Element for Modulating Biological Functions. Angew. Chem. Int. Ed. 2013, 52, 254–269. [Google Scholar] [CrossRef]
- Magafa, V.; Matsoukas, M.-T.; Karageorgos, V.; Dermitzaki, E.; Exarchakou, R.; Stylos, E.Κ.; Pardalos, M.; Margioris, A.N.; Varvounis, G.; Tzakos, A.G.; et al. Novel Stable Analogues of the Neurotensin C-Terminal Hexapeptide Containing Unnatural Amino Acids. Amino. Acids 2019, 51, 1009–1022. [Google Scholar] [CrossRef]
- Feni, L.; Jütten, L.; Parente, S.; Piarulli, U.; Neundorf, I.; Diaz, D. Cell-Penetrating Peptides Containing 2,5-Diketopiperazine (DKP) Scaffolds as Shuttles for Anti-Cancer Drugs: Conformational Studies and Biological Activity. Chem. Commun. 2020, 56, 5685–5688. [Google Scholar] [CrossRef]
- Pina, A.; Kadri, M.; Arosio, D.; Dal Corso, A.; Coll, J.; Gennari, C.; Boturyn, D. Multimeric Presentation of RGD Peptidomimetics Enhances Integrin Binding and Tumor Cell Uptake. Chem. Eur. J. 2020, 26, 7492–7496. [Google Scholar] [CrossRef]
- Formaggio, F.; Crisma, M.; Toniolo, C.; Tchertanov, L.; Guilhem, J.; Mazaleyrat, J.-P.; Gaucher, A.; Wakselman, M. Bip: A Ca-Tetrasubstituted, Axially Chiral a-Amino Acid. Synthesis and Conformational Preference of Model Peptides. Tetrahedron 2000, 56, 8721–8734. [Google Scholar] [CrossRef]
- Mazaleyrat, J.-P.; Gaucher, A.; Šavrda, J.; Wakselman, M. Novel α,α-Disubstituted α-Aminoacids with Axial Dissymmetry and Their N- or C-Protected Derivatives. Tetrahedron Asymmetry 1997, 8, 619–631. [Google Scholar] [CrossRef]
- Mazaleyrat, J.-P.; Gaucher, A.; Wakselman, M.; Tchertanov, L.; Guilhem, J. A New Chiral α-Aminoacid with Only Axial Dissymmetry: Synthesis and X-Ray Analysis of a 1,1′-Binaphthyl-Substituted α-Aminoisobutyric Acid (Bin) and of Its Biphenyl Analogue (Bip). Tetrahedron Lett. 1996, 37, 2971–2974. [Google Scholar] [CrossRef]
- Mazaleyrat, J.-P.; Wright, K.; Gaucher, A.; Toulemonde, N.; Wakselman, M.; Oancea, S.; Peggion, C.; Formaggio, F.; Setnička, V.; Keiderling, T.A.; et al. Induced Axial Chirality in the Biphenyl Core of the Cα-Tetrasubstituted α-Amino Acid Residue Bip and Subsequent Propagation of Chirality in (Bip)n/Val Oligopeptides. J. Am. Chem. Soc. 2004, 126, 12874–12879. [Google Scholar] [CrossRef]
- Dutot, L.; Wright, K.; Gaucher, A.; Wakselman, M.; Mazaleyrat, J.-P.; Zotti, M.D.; Peggion, C.; Formaggio, F.; Toniolo, C. The Bip Method, Based on the Induced Circular Dichroism of a Flexible Biphenyl Probe in Terminally Protected -Bip-Xaa*- Dipeptides, for Assignment of the Absolute Configuration of β-Amino Acids. J. Am. Chem. Soc. 2008, 130, 5986–5992. [Google Scholar] [CrossRef]
- Mazaleyrat, J.-P.; Wright, K.; Wakselman, M.; Formaggio, F.; Crisma, M.; Toniolo, C. 9-Amino-4,5-Diazafluorene-9-Carboxylic Acid (Daf), a New Cα,α-Disubstituted Glycine Containing a Spatially Constrained Bipyridine-Like Ligand for Transition Metals—Synthesis and Evaluation of Peptide-Coupling Conditions at ItsC- AndN-Termini. Eur. J. Org. Chem. 2001, 2001, 1821–1829. [Google Scholar] [CrossRef]
- Regan, L. The Design of Metal-Binding Sites in Proteins. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 257–281. [Google Scholar] [CrossRef]
- He, H.; Janso, J.E.; Yang, H.Y.; Bernan, V.S.; Lin, S.L.; Yu, K. Culicinin D, an Antitumor Peptaibol Produced by the Fungus Culicinomyces c Lavisporus, Strain LL -12I252. J. Nat. Prod. 2006, 69, 736–741. [Google Scholar] [CrossRef]
- Ko, K.-Y.; Wagner, S.; Yang, S.-H.; Furkert, D.P.; Brimble, M.A. Improved Synthesis of the Unnatural Amino Acids AHMOD and AMD, Components of the Anticancer Peptaibol Culicinin D. J. Org. Chem. 2015, 80, 8631–8636. [Google Scholar] [CrossRef]
- Lyu, P.C.; Sherman, J.C.; Chen, A.; Kallenbach, N.R. A-Helix Stabilization by Natural and Unnatural Amino Acids with Alkyl Side Chains. Proc. Natl. Acad. Sci. USA 1991, 88, 5317–5320. [Google Scholar] [CrossRef] [Green Version]
- Padmanabhan, S.; Baldwin, R.L. Straight-Chain Non-Polar Amino Acids Are Good Helix-Formers in Water. J. Mol. Biol. 1991, 219, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Soini, J.; Falschlehner, C.; Liedert, C.; Bernhardt, J.; Vuoristo, J.; Neubauer, P. Norvaline Is Accumulated after a Down-Shift of Oxygen in Escherichia Coli W3110. Microb. Cell Fact. 2008, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, X.-F.; Rajapakse, A.G.; Carvas, J.M.; Ruffieux, J.; Yang, Z. Inhibition of S6K1 Accounts Partially for the Anti-Inflammatory Effects of the Arginase Inhibitor L-Norvaline. BMC Cardiovasc. Disord. 2009, 9, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clementi, M.E.; Misiti, F. Substitution of Methionine 35 Inhibits Apoptotic Effects of Abeta(31-35) and Abeta(25-35) Fragments of Amyloid-Beta Protein in PC12 Cells. Med. Sci. Monit. 2005, 11, BR381–BR385. [Google Scholar] [PubMed]
- Andrews, M.J.I.; Tabor, A.B. Forming Stable Helical Peptides Using Natural and Artificial Amino Acids. Tetrahedron 1999, 55, 11711–11743. [Google Scholar] [CrossRef]
- Cornish, V.W.; Kaplan, M.I.; Veenstra, D.L.; Kollman, P.A.; Schultz, P.G. Stabilizing and Destabilizing Effects of Placing.Beta.-Branched Amino Acids in Protein.Alpha.-Helixes. Biochemistry 1994, 33, 12022–12031. [Google Scholar] [CrossRef]
- Jost, M.; Weigelt, S.; Huber, T.; Majer, Z.; Greie, J.-C.; Altendorf, K.; Sewald, N. Synthesis, and Structural and Biological Studies of Efrapeptin C Analogues. Chem. Biodivers. 2007, 4, 1170–1182. [Google Scholar] [CrossRef]
- Brückner, H.; Degenkolb, T. Sequences of Tolypins, Insecticidal Efrapeptin-Type Peptaibiotics from Species of the Fungal Genus Tolypocladium. Chem. Biodivers. 2020, 17, e2000276. [Google Scholar] [CrossRef]
- Wysong, C.L.; Yokum, T.S.; Morales, G.A.; Gundry, R.L.; McLaughlin, M.L.; Hammer, R.P. 4-Aminopiperidine-4-Carboxylic Acid: A Cyclic α,α-Disubstituted Amino Acid for Preparation of Water-Soluble Highly Helical Peptides. J. Org. Chem. 1996, 61, 7650–7651. [Google Scholar] [CrossRef]
- Toniolo, C. Structure of Conformationally Constrained Peptides: From Model Compounds to Bioactive Peptides. Biopolymers 1989, 28, 247–257. [Google Scholar] [CrossRef]
- Valle, G.; Crisma, M.; Toniolo, C.; Rao, R.B.; Sukumar, M.; Balaram, P. Stereochemistry of Peptides Containing 1-aminocycloheptane-1-carboxylic Acid (Ac7c). Int. J. Pept. Protein Res. 1991, 38, 511–518. [Google Scholar] [CrossRef]
- Rogers, J.M.; Suga, H. Discovering Functional, Non-Proteinogenic Amino Acid Containing, Peptides Using Genetic Code Reprogramming. Org. Biomol. Chem. 2015, 13, 9353–9363. [Google Scholar] [CrossRef]
- Rutjes, F.P.J.T.; Wolf, L.B.; Schoemaker, H.E. Applications of Aliphatic Unsaturated Non-Proteinogenic α-H-α-Amino Acids. J. Chem. Soc. Perkin Trans. 1 2000, 24, 4197–4212. [Google Scholar] [CrossRef]
- Guerrero, I.; Correa, A. Site-Selective Trifluoromethylation Reactions of Oligopeptides. Asian J. Org. Chem. 2020, 9, 898–909. [Google Scholar] [CrossRef]
- Bassetto, M.; Ferla, S.; Pertusati, F. Polyfluorinated Groups in Medicinal Chemistry. Future Med. Chem. 2015, 7, 527–546. [Google Scholar] [CrossRef]
- Wang, J.; Sánchez-Roselló, M.; Aceña, J.L.; del Pozo, C.; Sorochinsky, A.E.; Fustero, S.; Soloshonok, V.A.; Liu, H. Fluorine in Pharmaceutical Industry: Fluorine-Containing Drugs Introduced to the Market in the Last Decade (2001–2011). Chem. Rev. 2014, 114, 2432–2506. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, J.; Gu, Z.; Wang, S.; Zhu, W.; Aceña, J.L.; Soloshonok, V.A.; Izawa, K.; Liu, H. Next Generation of Fluorine-Containing Pharmaceuticals, Compounds Currently in Phase II-III Clinical Trials of Major Pharmaceutical Companies: New Structural Trends and Therapeutic Areas. Chem. Rev. 2016, 116, 422–518. [Google Scholar] [CrossRef]
- Ripka, A.S.; Rich, D.H. Peptidomimetic Design. Curr. Opin. Chem. Biol. 1998, 2, 441–452. [Google Scholar] [CrossRef]
- Cudic, P.; Stawikowski, M. Pseudopeptide Synthesis via Fmoc Solid-Phase Synthetic Methodology. Mini-Rev. Org. Chem. 2007, 4, 268–280. [Google Scholar] [CrossRef]
- Evans, B.J.; King, A.T.; Katsifis, A.; Matesic, L.; Jamie, J.F. Methods to Enhance the Metabolic Stability of Peptide-Based PET Radiopharmaceuticals. Molecules 2020, 25, 2314. [Google Scholar] [CrossRef]
- Ruzza, P. Peptides and Peptidomimetics in Medicinal Chemistry. In Medicinal Chemistry and Drug Design; Ekinci, D., Ed.; InTech: London, UK, 2012; ISBN 978-953-51-0513-8. [Google Scholar]
- Magrath, J.; Abeles, R.H. Cysteine Protease Inhibition by Azapeptide Esters. J. Med. Chem. 1992, 35, 4279–4283. [Google Scholar] [CrossRef]
- Tarchoun, K.; Yousef, M.; Bánóczi, Z. Azapeptides as an Efficient Tool to Improve the Activity of Biologically Effective Peptides. Future Pharmacol. 2022, 2, 293–305. [Google Scholar] [CrossRef]
- Chessum, N.; Jones, K.; Pasqua, E.; Tucker, M. Chapter One—Recent Advances in Cancer Therapeutics. In Progress in Medicinal Chemistry; Lawton, G., Witty, D.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; Volume 54, pp. 1–63. [Google Scholar]
- Sparidans, R.W.; Stokvis, E.; Jimeno, J.M.; López-Lázaro, L.; Schellens, J.H.; Beijnen, J.H. Chemical and Enzymatic Stability of a Cyclic Depsipeptide, the Novel, Marine-Derived, Anti-Cancer Agent Kahalalide F. Anti-Cancer Drugs 2001, 12, 575. [Google Scholar] [CrossRef]
- Farhid, H.; Rostami, M.M.; Shaabani, A.; Notash, B. Synthesis of Depsipeptides via Isocyanide-Based Consecutive Bargellini–Passerini Multicomponent Reactions. SynOpen 2021, 05, 167–172. [Google Scholar] [CrossRef]
- Hosono, Y.; Uchida, S.; Shinkai, M.; Townsend, C.E.; Kelly, C.N.; Naylor, M.R.; Lee, H.-W.; Kanamitsu, K.; Ishii, M.; Ueki, R.; et al. Amide-to-Ester Substitution as a Stable Alternative to N-Methylation for Increasing Membrane Permeability in Cyclic Peptides. Nat. Commun. 2023, 14, 1416. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, B.S.; Engh, R.A. Comparative Conformational Analyses and Molecular Dynamics Studies of Glycylglycine Methyl Ester and Glycylglycine N -Methylamide. RSC Adv. 2018, 8, 4445–4453. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Marine Natural Products and Related Compounds in Clinical and Advanced Preclinical Trials. J. Nat. Prod. 2004, 67, 1216–1238. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.M.; Holoye, P.Y.; Forman, A.; Winn, R.; Perez-Soler, R.; Dakhil, S.; Rosenthal, J.; Raber, M.N.; Hong, W.K. Phase II Clinical Trial of Didemnin B in Previously Treated Small Cell Lung Cancer. Investig. New Drugs 1994, 12, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Jimenez, G.-M.; Burgos-Hernandez, A.; Ezquerra-Brauer, J.-M. Bioactive Peptides and Depsipeptides with Anticancer Potential: Sources from Marine Animals. Mar. Drugs 2012, 10, 963–986. [Google Scholar] [CrossRef]
- Lee, Y.; Phat, C.; Hong, S.-C. Structural Diversity of Marine Cyclic Peptides and Their Molecular Mechanisms for Anticancer, Antibacterial, Antifungal, and Other Clinical Applications. Peptides 2017, 95, 94–105. [Google Scholar] [CrossRef]
- Mitsiades, C.S.; Ocio, E.M.; Pandiella, A.; Maiso, P.; Gajate, C.; Garayoa, M.; Vilanova, D.; Montero, J.C.; Mitsiades, N.; McMullan, C.J.; et al. Aplidin, a Marine Organism–Derived Compound with Potent Antimyeloma Activity In Vitro and In Vivo. Cancer Res. 2008, 68, 5216–5225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Pazgier, M.; Li, J.; Li, C.; Liu, M.; Zou, G.; Li, Z.; Chen, J.; Tarasov, S.G.; Lu, W.-Y.; et al. Limitations of Peptide Retro-Inverso Isomerization in Molecular Mimicry. J. Biol. Chem. 2010, 285, 19572–19581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arranz-Gibert, P.; Ciudad, S.; Seco, J.; García, J.; Giralt, E.; Teixidó, M. Immunosilencing Peptides by Stereochemical Inversion and Sequence Reversal: Retro-D-Peptides. Sci. Rep. 2018, 8, 6446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdini, A.S.; Silvestri, S.; Becherucci, C.; Longobardi, M.G.; Parente, L.; Peppoloni, S.; Perretti, M.; Pileri, P.; Pinori, M. Immunostimulation by a Partially Modified Retro-Inverso-Tuftsin Analog Containing Thr1.Sum..Psi.[NHCO](R,S)Lys2 Modification. J. Med. Chem. 1991, 34, 3372–3379. [Google Scholar] [CrossRef] [PubMed]
- Doti, N.; Mardirossian, M.; Sandomenico, A.; Ruvo, M.; Caporale, A. Recent Applications of Retro-Inverso Peptides. Int. J. Mol. Sci. 2021, 22, 8677. [Google Scholar] [CrossRef]
- Preston, G.W. Different Directions for Retro-Inverso Peptides. J. Pept. Sci. 2022, 28, e3384. [Google Scholar] [CrossRef]
- Rai, J. Peptide and Protein Mimetics by Retro and Retroinverso Analogs. Chem. Biol. Drug Des. 2019, 93, 724–736. [Google Scholar] [CrossRef]
- Simon, R.J.; Kania, R.S.; Zuckermann, R.N.; Huebner, V.D.; Jewell, D.A.; Banville, S.; Ng, S.; Wang, L.; Rosenberg, S.; Marlowe, C.K. Peptoids: A Modular Approach to Drug Discovery. Proc. Natl. Acad. Sci. USA 1992, 89, 9367–9371. [Google Scholar] [CrossRef] [Green Version]
- Kirshenbaum, K.; Barron, A.E.; Goldsmith, R.A.; Armand, P.; Bradley, E.K.; Truong, K.T.V.; Dill, K.A.; Cohen, F.E.; Zuckermann, R.N. Sequence-Specific Polypeptoids: A Diverse Family of Heteropolymers with Stable Secondary Structure. Proc. Natl. Acad. Sci. USA 1998, 95, 4303–4308. [Google Scholar] [CrossRef] [Green Version]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids That Mimic the Structure, Function, and Mechanism of Helical Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef] [Green Version]
- Edison, J.R.; Spencer, R.K.; Butterfoss, G.L.; Hudson, B.C.; Hochbaum, A.I.; Paravastu, A.K.; Zuckermann, R.N.; Whitelam, S. Conformations of Peptoids in Nanosheets Result from the Interplay of Backbone Energetics and Intermolecular Interactions. Proc. Natl. Acad. Sci. USA 2018, 115, 5647–5651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, A.; Baer, M.D.; Mundy, C.J.; Pfaendtner, J. Peptoid Backbone Flexibilility Dictates Its Interaction with Water and Surfaces: A Molecular Dynamics Investigation. Biomacromolecules 2018, 19, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Wellhöfer, I.; Frydenvang, K.; Kotesova, S.; Christiansen, A.M.; Laursen, J.S.; Olsen, C.A. Functionalized Helical β-Peptoids. J. Org. Chem. 2019, 84, 3762–3779. [Google Scholar] [CrossRef] [PubMed]
- Kann, N.; Johansson, J.R.; Beke-Somfai, T. Conformational Properties of 1,4- and 1,5-Substituted 1,2,3-Triazole Amino Acids—Building Units for Peptidic Foldamers. Org. Biomol. Chem. 2015, 13, 2776–2785. [Google Scholar] [CrossRef] [Green Version]
- Staśkiewicz, A.; Ledwoń, P.; Rovero, P.; Papini, A.M.; Latajka, R. Triazole-Modified Peptidomimetics: An Opportunity for Drug Discovery and Development. Front. Chem. 2021, 9, 674705. [Google Scholar] [CrossRef]
- Martinek, T.A.; Fülöp, F. Peptidic Foldamers: Ramping up Diversity. Chem. Soc. Rev. 2012, 41, 687–702. [Google Scholar] [CrossRef]
- Lesma, G.; Salvadori, S.; Airaghi, F.; Murray, T.F.; Recca, T.; Sacchetti, A.; Balboni, G.; Silvani, A. Structural and Biological Exploration of Phe 3 –Phe 4 -Modified Endomorphin-2 Peptidomimetics. ACS Med. Chem. Lett. 2013, 4, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Gavras, H.; Brunner, H.R. Role of Angiotensin and Its Inhibition in Hypertension, Ischemic Heart Disease, and Heart Failure. Hypertension 2001, 37, 342–345. [Google Scholar] [CrossRef] [Green Version]
- Bork, K.; Yasothan, U.; Kirkpatrick, P. Icatibant. Nat. Rev. Drug Discov. 2008, 7, 801–802. [Google Scholar] [CrossRef]
- Gruber, C.W.; Koehbach, J.; Muttenthaler, M. Exploring Bioactive Peptides from Natural Sources for Oxytocin and Vasopressin Drug Discovery. Future Med. Chem. 2012, 4, 10–4155. [Google Scholar] [CrossRef] [Green Version]













| ncAA Class | Highlights | Conformational Preferences | Characteristics | Application |
|---|---|---|---|---|
| symmetric α,α-dialkyl glycines | Aib | 310-helix or α-helix | increased proteolytic resistance helical foldamers | antimicrobial/antibiotic peptidomimetics |
| Dhg | α-helix | |||
| asymmetric α,α-dialkyl glycines (d-amino acids) | Iva, MDL and MDP | 310-helix or α-helix | increased proteolytic resistance helical foldamers | antimicrobial/antibiotic peptidomimetics |
| Cα to Cα cyclized (Acnc residues) | Ac3c | bridge region | foldamers | neurotransmitters antimicrobial/antibiotic peptidomimetics |
| Ac6c | 310-helix or α-helix | |||
| (R,R)Ac5cdOM | 310-helix or α-helix | |||
| proline analogues | Hyp | β-turn, bend | foldamers | antimicrobial peptidomimetics |
| β-substituted amino acids | β-MePhe Tmt, Tic | Side-chain constraint | increased proteolytic resistance hormones mimetics | antinociceptive activity (opioids) |
| α,β-dehydroamino acids | ΔzPhe ΔAbu | β-turn or γ-turn 310-helix or α-helix | increased proteolytic resistance hydrogels | drug delivery cancer treatment |
| N-alkylated | sarcosine | cyclic peptides | increased lipophilicity improved pharmacokinetics | antibiotic immunosuppressant |
| N-cyclization | DKP1 DKP3 | helix | increased proteolytic resistance | neurotransmitter neuromodulator drug delivery anticancer |
| other | Bip Bin | turn/helix inducers | increased proteolytic resistance | circular dichroism probe |
| other | (S)-Ethynylglycine | β-turn β-hairpin | foldamers | antibiotic activity |
| Backbone Modification | Highlights | Conformational Preferences | Characteristics | Application |
|---|---|---|---|---|
| azapeptides | Ac-l-Phe-azaAlaOiB Ac-l-Phe-azaGlyOMe Boc-(Phe-azaPhe-Ala)2-OMe) | β-turn extended | increased proteolytic stability | inhibitors of cysteine proteases |
| depsipeptides | Didemnin B Plitidepsin Kahalalide F Romidepsin | cyclic | increased flexibility | antiviral cancers treatments |
| retro-inverso | Amytrap BMAP-28D(LPR); D(RGD) | extended helix | resistant to proteolytic degradation | anticancer immunology neurodegenerative diseases antimicrobial diagnosis |
| peptoids | Triazole-peptidomimetics | helix sheets | stable synthetic polymers | antimicrobial drug carrier anticancer antibiotics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, T.G.; Melle-Franco, M.; Sousa, C.E.A.; Cavaco-Paulo, A.; Marcos, J.C. Non-Canonical Amino Acids as Building Blocks for Peptidomimetics: Structure, Function, and Applications. Biomolecules 2023, 13, 981. https://doi.org/10.3390/biom13060981
Castro TG, Melle-Franco M, Sousa CEA, Cavaco-Paulo A, Marcos JC. Non-Canonical Amino Acids as Building Blocks for Peptidomimetics: Structure, Function, and Applications. Biomolecules. 2023; 13(6):981. https://doi.org/10.3390/biom13060981
Chicago/Turabian StyleCastro, Tarsila G., Manuel Melle-Franco, Cristina E. A. Sousa, Artur Cavaco-Paulo, and João C. Marcos. 2023. "Non-Canonical Amino Acids as Building Blocks for Peptidomimetics: Structure, Function, and Applications" Biomolecules 13, no. 6: 981. https://doi.org/10.3390/biom13060981