Selenium-Substituted Monomethine Cyanine Dyes as Selective G-Quadruplex Spectroscopic Probes with Theranostic Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Synthetic Methods






2.2. Materials and Spectrophotometric Methods
2.3. Effect of Compounds on Cell Viability
2.3.1. Cell Lines
2.3.2. Cytotoxicity Assay
2.4. Co-Localization Assay
3. Results and Discussion of Spectrophotometric Study
3.1. Characterization of Compounds in Aqueous Medium
3.2. Study of Interactions of 1–6 with Nucleic Acids in Aqueous Medium
3.2.1. Interactions with ds-DNA and ds-RNA
3.2.2. Interactions with G-Quadruplex
4. Biological Activity
4.1. MTT Assay
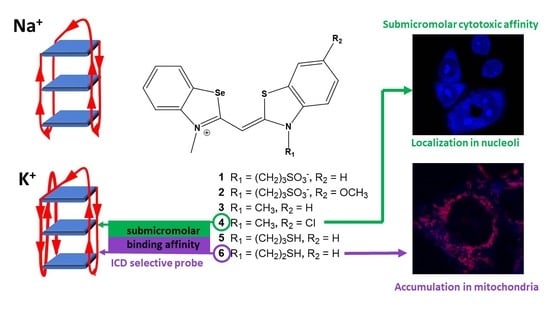
4.2. Confocal Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Demeunynck, M.; Bailly, C.; Wilson, W.D. Small Molecule DNA and RNA Binders; Wiley-VCH: Weinheim, Germany, 2002. [Google Scholar]
- Zonjic, I.; Tumir, L.M.; Crnolatac, I.; Supljika, F.; Racane, L.; Tomic, S.; Radic Stojkovic, M. Recognition of ATT Triplex and DNA:RNA Hybrid Structures by Benzothiazole Ligands. Biomolecules 2022, 12, 374. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Oxford Handbook of Nucleic acid Structure; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Armitage, B.A. Cyanine dye-DNA interactions: Intercalation, groove binding, and aggregation. Top. Curr. Chem. 2005, 253, 55–76. [Google Scholar]
- Yarmoluk, S.M.; Lukashov, S.S.; Ogul’Chansky, T.Y.; Losytskyy, M.Y.; Kornyushyna, O.S. Interaction of cyanine dyes with nucleic acids. XXI. Arguments for half-intercalation model of interaction. Biopolymers 2001, 62, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Biancardi, A.; Biver, T.; Marini, A.; Mennucci, B.; Secco, F. Thiazole orange (TO) as a light-switch probe: A combined quantum-mechanical and spectroscopic study. Phys. Chem. Chem. Phys. 2011, 13, 12595–12602. [Google Scholar] [CrossRef] [PubMed]
- Kandinska, M.I.; Cheshmedzhieva, D.V.; Kostadinov, A.; Rusinov, K.; Rangelov, M.; Todorova, N.; Ilieva, S.; Ivanov, D.P.; Videva, V.; Lozanov, V.S.; et al. Tricationic asymmetric monomeric monomethine cyanine dyes with chlorine and trifluoromethyl functionality–Fluorogenic nucleic acids probes. J. Mol. Liq. 2021, 342, 117501. [Google Scholar] [CrossRef]
- Glavas-Obrovac, L.; Piantanida, I.; Marczi, S.; Masic, L.; Timcheva, I.I.; Deligeorgiev, T.G. Minor structural differences of monomethine cyanine derivatives yield strong variation in their interactions with DNA, RNA as well as on their in vitro antiproliferative activity. Bioorg. Med. Chem. 2009, 17, 4747–4755. [Google Scholar] [CrossRef] [PubMed]
- Tumir, L.M.; Crnolatac, I.; Deligeorgiev, T.; Vasilev, A.; Kaloyanova, S.; Grabar Branilovic, M.; Tomic, S.; Piantanida, I. Kinetic differentiation between homo- and alternating AT DNA by sterically restricted phosphonium dyes. Chemistry 2012, 18, 3859–3864. [Google Scholar] [CrossRef]
- Smidlehner, T.; Koscak, M.; Bozinovic, K.; Majhen, D.; Schmuck, C.; Piantanida, I. Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate. Molecules 2020, 25, 4470. [Google Scholar] [CrossRef]
- Rožman, A.; Crnolatac, I.; Deligeorgiev, T.; Piantanida, I. Strong impact of chloro substituent on TOTO and YOYO ds-DNA/RNA sensing. J. Lumin. 2019, 205, 87–96. [Google Scholar] [CrossRef]
- Mikulin, I.; Ljubic, I.; Piantanida, I.; Vasilev, A.; Mondeshki, M.; Kandinska, M.; Uzelac, L.; Martin-Kleiner, I.; Kralj, M.; Tumir, L.M. Polycationic Monomeric and Homodimeric Asymmetric Monomethine Cyanine Dyes with Hydroxypropyl Functionality-Strong Affinity Nucleic Acids Binders. Biomolecules 2021, 11, 1075. [Google Scholar] [CrossRef]
- Zonjic, I.; Radic Stojkovic, M.; Crnolatac, I.; Tomasic Paic, A.; Psenicnik, S.; Vasilev, A.; Kandinska, M.; Mondeshki, M.; Baluschev, S.; Landfester, K.; et al. Styryl dyes with N-Methylpiperazine and N-Phenylpiperazine Functionality: AT-DNA and G-quadruplex binding ligands and theranostic agents. Bioorg. Chem. 2022, 127, 105999. [Google Scholar] [CrossRef] [PubMed]
- Zonjić, I.; Kurutos, A.; Mihovilović, P.; Crnolatac, I.; Tumir, L.M.; Tomašić Paić, A.; Kralj, J.; Horvat, L.; Brozovic, A.; Stojković, R.; et al. Formation of triplex rA/dA-containing nucleic acid helices induced by new thiazole orange analogues and their biological evaluation. H-aggregate formation conditioned by rA sequence. Dye. Pigment. 2022, 207, 110715. [Google Scholar] [CrossRef]
- Sun, J.; Feng, E.; Shao, Y.; Lv, F.; Wu, Y.; Tian, J.; Sun, H.; Song, F. A Selenium-Substituted Heptamethine Cyanine Photosensitizer for Near-Infrared Photodynamic Therapy. ChemBioChem 2022, 23, e202200421. [Google Scholar] [CrossRef]
- Xu, K.; Chen, H.; Tian, J.; Ding, B.; Xie, Y.; Qiang, M.; Tang, B. A near-infrared reversible fluorescent probe for peroxynitrite and imaging of redox cycles in living cells. Chem. Commun. 2011, 47, 9468–9470. [Google Scholar] [CrossRef] [PubMed]
- Kurutos, A.; Ryzhova, O.; Trusova, V.; Gorbenko, G.; Gadjev, N.; Deligeorgiev, T. Symmetric Meso-Chloro-Substituted Pentamethine Cyanine Dyes Containing Benzothiazolyl/Benzoselenazolyl Chromophores Novel Synthetic Approach and Studies on Photophysical Properties upon Interaction with bio-Objects. J. Fluoresc. 2016, 26, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.L.; Maia, A.; Santos, A.O.; Lima, E.; Reis, L.V.; Nunes, M.J.; Boto, R.E.F.; Silvestre, S.; Almeida, P. An Insight into Symmetrical Cyanine Dyes as Promising Selective Antiproliferative Agents in Caco-2 Colorectal Cancer Cells. Molecules 2022, 27, 5779. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.C.; Wu, T.Y.; Zeng, S.T.; Fang, L.; Mao, J.X.; Chen, S.B.; Huang, Z.S.; Chen, X.C.; Tan, J.H. Benzoselenazolium-based hemicyanine dye for G-Quadruplex detection. Bioorg. Med. Chem. Lett. 2022, 70, 128801. [Google Scholar] [CrossRef]
- Yan, J.; Guo, Y.; Wang, Y.; Mao, F.; Huang, L.; Li, X. Design, synthesis, and biological evaluation of benzoselenazole-stilbene hybrids as multi-target-directed anti-cancer agents. Eur. J. Med. Chem. 2015, 95, 220–229. [Google Scholar] [CrossRef]
- Kremer, A.; Fermi, A.; Biot, N.; Wouters, J.; Bonifazi, D. Supramolecular Wiring of Benzo-1,3-chalcogenazoles through Programmed Chalcogen Bonding Interactions. Chemistry 2016, 22, 5665–5675. [Google Scholar] [CrossRef]
- Reis, L.V.; Serrano, J.P.C.; Almeida, P.; Santos, P.F. New synthetic approach to aminosquarylium cyanine dyes. Synlett 2002, 10, 1617–1620. [Google Scholar]
- Kurutos, A.; Gadjev, N.; Deligeorgiev, T. Synthesis of novel asymmetric monomethine cyanine dyes containing benzoselenazolyl moiety. Fluorescent dsDNA probes. Acta Sci. Nat. 2015, 90–98. [Google Scholar]
- Chaires, J.B.; Dattagupta, N.; Crothers, D.M. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: Equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982, 21, 3933–3940. [Google Scholar] [CrossRef] [PubMed]
- Bresloff, J.L.; Crothers, D.M. Equilibrium studies of ethidium–polynucleotide interactions. Biochemistry 1981, 20, 3547–3553. [Google Scholar] [CrossRef] [PubMed]
- Chalikian, T.V.; Volker, J.; Plum, G.E.; Breslauer, K.J. A more unified picture for the thermodynamics of nucleic acid duplex melting: A characterization by calorimetric and volumetric techniques. Proc. Natl. Acad. Sci. USA 1999, 96, 7853–7858. [Google Scholar] [CrossRef] [Green Version]
- Scatchard, G. The Attractions of Proteins for Small Molecules and Ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef]
- Mickisch, G.; Fajta, S.; Keilhauer, G.; Schlick, E.; Tschada, R.; Alken, P. Chemosensitivity testing of primary human renal cell carcinoma by a tetrazolium based microculture assay (MTT). Urol. Res. 1990, 18, 131–136. [Google Scholar] [CrossRef]
- Furstenberg, A.; Julliard, M.D.; Deligeorgiev, T.G.; Gadjev, N.I.; Vasilev, A.A.; Vauthey, E. Ultrafast excited-state dynamics of DNA fluorescent intercalators: New insight into the fluorescence enhancement mechanism. J. Am. Chem. Soc. 2006, 128, 7661–7669. [Google Scholar] [CrossRef] [Green Version]
- Saenger, W. Principles of Nucleic acid Structure; Springer-Verlag: New York, NY, USA, 1984. [Google Scholar]
- Cantor, C.R. Techniques for the Study of Biological Structure and Function; W. H. Freeman: San Francisco, CA, USA, 1980. [Google Scholar]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Wilson, W.D.; Ratmeyer, L.; Zhao, M.; Strekowski, L.; Boykin, D. The search for structure-specific nucleic acid-interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry 1993, 32, 4098–4104. [Google Scholar] [CrossRef] [PubMed]
- Tumir, L.M.; Zonjic, I.; Zuna, K.; Brkanac, S.R.; Jukic, M.; Hudek, A.; Durgo, K.; Crnolatac, I.; Glavas-Obrovac, L.; Cardullo, N.; et al. Synthesis, DNA/RNA-interaction and biological activity of benzo[k,l]xanthene lignans. Bioorg. Chem. 2020, 104, 104190. [Google Scholar] [CrossRef] [PubMed]
- Rodger, A.; Nordén, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997; Chapter 2. [Google Scholar]
- Eriksson, M.; Norden, B. Linear and circular dichroism of drug-nucleic acid complexes. Methods Enzym. 2001, 340, 68–98. [Google Scholar]
- Smidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids–tutorial. Beilstein J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Chaires, J.B. Human telomeric G-quadruplex: Thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010, 277, 1098–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cree, S.L.; Kennedy, M.A. Relevance of G-quadruplex structures to pharmacogenetics. Front. Pharm. 2014, 5, 160. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef] [Green Version]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [Green Version]
- Gampp, H.; Maeder, M.; Meyer, C.J.; Zuberbuhler, A.D. Calculation of equilibrium constants from multiwavelength spectroscopic data–II: SPECFIT: Two user-friendly programs in basic and standard FORTRAN 77. Talanta 1985, 32, 257–264. [Google Scholar] [CrossRef]
- Chen, S.B.; Tan, J.H.; Ou, T.M.; Huang, S.L.; An, L.K.; Luo, H.B.; Li, D.; Gu, L.Q.; Huang, Z.S. Pharmacophore-based discovery of triaryl-substituted imidazole as new telomeric G-quadruplex ligand. Bioorg. Med. Chem. Lett. 2011, 21, 1004–1009. [Google Scholar] [CrossRef]
- Zhou, J.; Le, V.; Kalia, D.; Nakayama, S.; Mikek, C.; Lewis, E.A.; Sintim, H.O. Diminazene or berenil, a classic duplex minor groove binder, binds to G-quadruplexes with low nanomolar dissociation constants and the amidine groups are also critical for G-quadruplex binding. Mol. Biosyst. 2014, 10, 2724–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvlin, M.J.; Datta-Gupta, N.; Fiel, R.J. Circular dichroism spectroscopy of a cationic porphyrin bound to DNA. Biochem Biophys Res. Commun. 1982, 108, 66–73. [Google Scholar] [CrossRef]
- Ramos, C.I.V.; Almeida, S.P.; Lourenco, L.M.O.; Pereira, P.M.R.; Fernandes, R.; Faustino, M.A.F.; Tome, J.P.C.; Carvalho, J.; Cruz, C.; Neves, M. Multicharged Phthalocyanines as Selective Ligands for G-Quadruplex DNA Structures. Molecules 2019, 24, 733. [Google Scholar] [CrossRef] [Green Version]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef]
- Crnolatac, I. Getting charged for the mitochondrial uptake. In Fluorescence Methods for Investigation of Living Cells and Microorganisms; Grigoryeva, N., Ed.; IntechOpen Limited: London, UK, 2020; pp. 1–25. [Google Scholar]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Adler, J.; Pagakis, S.N.; Parmryd, I. Replicate-based noise corrected correlation for accurate measurements of colocalization. J. Microsc. 2008, 230, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.; Chen, H.; Li, Q.; Guan, A.; Wang, L.; Shi, Y.; Xu, S.; Liu, M.; Tang, Y. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1101–1106. [Google Scholar] [CrossRef]
- Falabella, M.; Fernandez, R.J.; Johnson, F.B.; Kaufman, B.A. Potential Roles for G-Quadruplexes in Mitochondria. Curr. Med. Chem. 2019, 26, 2918–2932. [Google Scholar] [CrossRef] [PubMed]









| Compd | UV/Vis λmax (nm) | ε × 103/mmol−1 cm2 |
|---|---|---|
| 1 | 425 | 76.7 |
| 2 | 431 | 37.6 |
| 3 | 423 | 69.7 |
| 4 | 427 | 55.2 |
| 5 | 406 | 42.0 |
| 6 | 407 | 30.6 |
| Compound | logKa1; logKa2 b | ∆Tm/°C |
|---|---|---|
| 1 | 6.93 ± 0.25; 5.69 ± 0.23 | 0 |
| 2 | - c | 0 |
| 3 | 7.26 ± 0.41; 5.93 ± 0.47 | 0.5 |
| 4 | 6.19 ± 0.01 | 2.1 |
| 5 | - c | 2.1 |
| 6 | - c | 2.0 |
| Compound | logKa b | ∆Tm/°C |
|---|---|---|
| 1 | - c | - d |
| 2 | - d | - d |
| 3 | 6.88 ± 0.05 | 0.5 |
| 4 | 7.78 ± 0.18 | 3.2 |
| 5 | - c | 5.1 |
| 6 | - c | 6.9 |
| 1 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|
| HeLa | 31.856 ± 1.142 | 1.025 ± 0.672 | 0.793 ± 0.157 | 27.917 ± 4.141 | 30.667 ± 4.072 |
| MDA-MB-435S | 18.525 ± 1.226 | 0.987 ± 0.177 | 0.6135 ± 0.0776 | 37.115 ± 3.021 | 32.767 ± 3.756 |
| SK-OV-3 | 32.75 ± 1.09 | 0.548 ± 0.102 | 1.319 ± 0.125 | 29.5625 ± 5.724 | >40 |
| Fibroblasts | >40 | 2.375 ± 0.161 | >2.5 | >40 | >40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabijanić, I.; Kurutos, A.; Tomašić Paić, A.; Tadić, V.; Kamounah, F.S.; Horvat, L.; Brozovic, A.; Crnolatac, I.; Radić Stojković, M. Selenium-Substituted Monomethine Cyanine Dyes as Selective G-Quadruplex Spectroscopic Probes with Theranostic Potential. Biomolecules 2023, 13, 128. https://doi.org/10.3390/biom13010128
Fabijanić I, Kurutos A, Tomašić Paić A, Tadić V, Kamounah FS, Horvat L, Brozovic A, Crnolatac I, Radić Stojković M. Selenium-Substituted Monomethine Cyanine Dyes as Selective G-Quadruplex Spectroscopic Probes with Theranostic Potential. Biomolecules. 2023; 13(1):128. https://doi.org/10.3390/biom13010128
Chicago/Turabian StyleFabijanić, Ivana, Atanas Kurutos, Ana Tomašić Paić, Vanja Tadić, Fadhil S. Kamounah, Lucija Horvat, Anamaria Brozovic, Ivo Crnolatac, and Marijana Radić Stojković. 2023. "Selenium-Substituted Monomethine Cyanine Dyes as Selective G-Quadruplex Spectroscopic Probes with Theranostic Potential" Biomolecules 13, no. 1: 128. https://doi.org/10.3390/biom13010128