1. Introduction
Homologous DNA recombination includes DNA strand exchange and occurs in all forms of life in the context of DNA repair and genetic stability [
1]. Various reports show that in living organisms, recombination enzymes, such as RecA in prokaryotes or its homolog Rad51 in eukaryotes, catalyze the process of strand displacement [
2,
3]. Although the details of this enzymatically mediated strand displacement process of duplex DNA are only partially understood, it is proposed that the DNA strand exchange involves at least three sequential steps. First, a single strand (ss) DNA diffuses in proximity of the double-strand (ds) DNA; second, the displacement strand interacts with one strand of the double-strand DNA and leads to an exchange of base pairing of a few bases; third, the strand is exchanged along the DNA duplex [
2,
3].
DNA strand displacement is becoming an essential process in nanotechnology applications as well. DNA is recognized as material for nanoscale engineering due to its specificity and programmability controlled by Watson-Crick canonical base pairing [
4]. These features allow the use of DNA for the construction of extremely precise nanostructures through the programmed hybridization of complementary strands [
5,
6]. Initially, DNA-based nanotechnology focused on the self-assembly of static structures. Subsequently, the field blossomed with the development of the DNA origami method, empowering the design and generation of “dynamic” structures. The function of these structures can be controlled and modulated by conformational changes induced by external stimuli or strand-displacement reactions [
7]. For instance, dynamic DNA structures, which are based on a toehold-mediated strand displacement, [
8] are used in different applications, including artificial molecular machines and programmable scaffolds [
9]. In particular, the approach of toehold-mediated strand displacement is based on the attachment of an invader strand to the toehold present on the duplex, followed by stepwise dislocation of one strand of the substrate duplex until full hybridization of the complementary invader to the substrate strand is completed [
10,
11].
There is a need for a better understanding of the strand displacement process to reveal further fundamental mechanisms of these interactions, which are potentially important in vivo. In addition, controlled manipulation of the strand displacement process in vitro can increase the rate of the displacement reaction and the yield of assembled nanostructures, which will accelerate further applications in nanotechnology. Feng et al. reported increased DNA strand exchange rates in concentrated aqueous solutions of polyethylene glycol (peg) [
12]. In the following study, the same authors highlighted that base stacking destabilization and nucleation-promoted DNA strand invasion are two core mechanisms that contribute to the catalytic role of the two semi-hydrophobic crowders used, peg and 1,2-dimethoxyethane (dme) [
13]. Very recently, Norden et al. used a mismatched duplex on different positions and showed that the mismatch is displaced faster into a matched duplex with a matched DNA strand if a peg is present as a crowder [
14]. They concluded that the hydrophobic interactions between the peg and the oligonucleotides are likely the cause of this catalytic effect and excluded molecular crowding as a possible mechanism.
Herein, we investigated and compared, for the first time, the strand exchange kinetics and the base-pairing fidelity of natural DNA, natural RNA, and LNA-modified DNA sequences as displacement strands. We included diethylene glycol dimethyl ether (deg), which could help solubilize DNA via hydrophobic interactions without exerting a crowding effect due to the small size, and the macromolecular crowders peg and polyvinylpyrrolidone (pvp) (
Figure 1) to model crowding inside living cells.
Our results indicate that the different chemical natures of the strands, including DNA, RNA, or LNA-DNA, have distinct displacement kinetics in pure PBS, with the LNA-DNA showing the fastest displacement. The different buffer additives used further affected the kinetics to variable degrees. Overall, we show that concerning the strand displacement kinetics, interactions of the additives with the hybridized strands have a big effect that can exceed the effect exerted by the nature of the displacement strand itself.
2. Materials and Methods
Reagents and solvents were obtained from commercial suppliers and used without further purification. The oligonucleotide sequences were supplied by Biomers (Ulm, Germany) and used as received. Polyethylene glycol (peg_6000), diethylene glycol dimethyl ether (deg), and polyvinylpyrrolidone (pvp_360,000) were purchased from Sigma (Buchs, Switzerland). We annealed the final master mix using an oligonucleotide concentration of 1 µM for each strand in the total volume of 100 µL.
2.1. Sample Preparation and Fluorescence Measurements
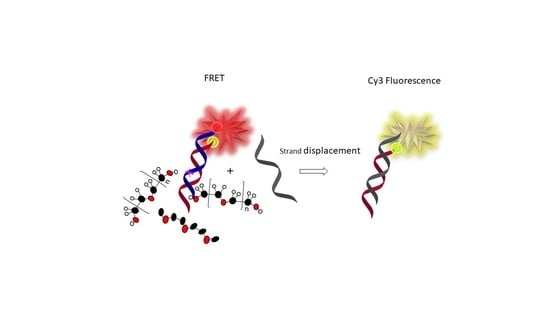
Annealing of the FRET pair duplex oligonucleotides (Bmut-CoBmut, 1000 nM in a 100 µL) was performed by exposing the mixture to 90 °C for 10 min and cooling down at room temperature for 2 h, followed by chilling at +4 °C for 1 h. Once the duplex Bmut-CoBmut was formed, it exhibited FRET upon excitation. Next, the crowders, peg 45%, deg 45%, and pvp 10% (concentrations given as w/w) were introduced for a minimum time of 15 min. Immediately after the addition of the displacement strand (DS1, DS2, DS3, or DSCtrl, in a five-fold molar excess), the Cy5 fluorescence emission over time was measured on a BioTek Cytation 5 Imaging Multi-Mode reader for a minimum of 30 min. To monitor FRET, we excited the donor Cy3 dye at 535 nm and followed the fluorescence emission of the acceptor Cy5 at 675 nm. We measured the kinetics in a 37 °C temperature-controlled instrument. As the displacement reaction proceeds, the FRET pair dissociates, resulting in a decrease in the FRET efficiency between Cy3 and Cy5, and decreased Cy5 fluorescence emission.
2.2. Circular Dichroism (CD) Measurements
CD measurements were performed on a Chirascan CD spectropolarimeter. The ds (2 µM from each strand) was annealed as described above, and the crowders were introduced for a minimum time of 15 min before the measurements. The CD measurements were performed at 37 °C using bandwidth of 1 nm and time per point of 1 s.
2.3. Viscosity Measurements
Viscosity measurements were performed with a 50 mm diameter cone and plate geometry mounted on a stress-controlled rheometer (MCR300, Paar Physica, Anton Paar, Virginia, NB, USA). The sample temperature of 37 °C was maintained constant by a Peltier module. For each solution, the value of the viscosity was obtained from the ratio between the measured stress and the imposed shear rate for shear rates in the range of 10–1000 1/s. As the viscosity is independent on the shear rate for each solution, we reported the mean and standard deviations for different shear rates. The kinetic constants determined from the data fitting were multiplied by the buffer viscosity (
Table S1) to calculate the viscosity-independent constants reported in
Figure S4. In fact, it is known from collision theory that the kinetic constant of diffusion-governed bimolecular processes depends on the inverse of solvent viscosity [
15].
2.4. Kinetic Modelling and Curve Fitting
A simple kinetic model describing an irreversible displacement reaction was used to analyze the experimental data:
In Equation (1),
indicates the original duplex, while
is the displacement strand and
is the displacement constant with units L·mol
−1·min
−1. This reaction scheme can be described by the following rate equation:
where the brackets indicate concentrations and
is time. If
is known, Equation (2) can be numerically integrated under the constraints that
µM,
µM,
,
, corresponding to our initial experimental conditions. However,
is unknown. To determine
, we, therefore, embedded the numerical integration routine into a fitting algorithm. The fitting algorithm uses an initial guess for the value of
to numerically determine the value of
as a function of time. The obtained
curve is then normalized with respect to
and compared to the normalized fluorescence intensity
under the assumption that the measured fluorescence intensity is proportional to
. The procedure is repeated iteratively by varying
until the
value that minimizes the difference between numerically predicted and experimental data is found.
The fitting algorithm was based on Python’s LMFIT (
https://lmfit.github.io/lmfit-py/, 3 September 2022) package using a nonlinear least squares method (‘leastsq’ method in scipy). The numerical integration of the displacement rate equation was performed using the scipy.integrate solve_ivp module (
https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.solve_ivp.html, 3 September 2022) with the explicit Runge-Kutta method of order 8 (‘DOP853’). The same procedure has been used to test more comprehensive kinetic models comprising multiple reversible steps. However, increasing the number of fitting parameters quickly resulted in overfitting, fitting instabilities, and a strong dependence of the fitting results from the initial guesses. Hence, only the results obtained with the single-parameter model (Equations (1) and (2)) were deemed reliable and are reported in this work.
4. Discussion
In this work, we studied the strand exchange kinetics of three different oligonucleotides: DNA (DS1), RNA (DS2), and LNA modified DNA sequence (DS3) in various buffers with or without molecular and macromolecular crowders of different sizes to simulate a semi-hydrophobic (peg and deg) environment in vitro. In addition to strand exchange phenomena between double-stranded and single-stranded DNA occurring naturally during homologous DNA recombination, the CRISPR-Cas-based gene editing machinery also involves RNA-DNA recognition and duplex formation [
21]. Moreover, many LNA-containing DNA sequences are employed in drug discovery studies, such as oligonucleotide therapeutics [
22] and advanced modified sgRNA for CRISPR-Cas [
23].
Our data indicate that the kinetic strand displacement depends on the nature of the newly introduced strand and on the nature of the crowders. In general, comparing the three displacement strands, the displacement capacity of DS3 was the least affected by the different buffer conditions. More specifically, in a pure PBS, the LNA-DNA oligonucleotide (DS3) can convert the mismatched duplex into a matched one fastest and to a greater extent compared with the natural DNA (DS1) and RNA (DS2) sequence. These results suggest that when a DNA strand contains LNA modifications, its base-pairing fidelity and strand displacement kinetics increase. These findings are consistent with previously reported properties of the LNA modification introduced to a DNA sequence to increase the target binding specificity [
24,
25,
26].
However, in the peg-containing buffer, DS1 and DS3 showed similar and faster displacement kinetics compared to DS2. In this buffer condition, while the displacement kinetics of DS1 was slightly increased, it was slightly decreased for DS2 and DS3 when compared with the PBS condition. Nordèn et al. previously reported that a matched DNA strand could displace a mismatch strand in a duplex faster in a solution containing non-ionic semi-hydrophobic peg molecules [
14]. The authors excluded that the cause for this effect is peg-induced variations in the DNA duplex conformation or duplex melting. Instead, they proposed a hydrophobic effect mechanism that includes three different steps. First, interruption of the base stacking and strand invasion; second, stronger hydrogen bonds by decreased water activity; and third, stabilization of the mismatch by intercalation of the hydrophobic crowder into the mismatched gaps [
14]. In agreement with this report, we confirm slightly faster displacement kinetics of DNA displacement strands in a peg-buffer. Nevertheless, the additional data on DS2 and DS3 displacement strands reveal that peg-enriched buffer fails to increase the displacement kinetics of RNA and LNA-DNA displacement strands.
Notably, the data from the experiments performed in a buffer containing deg reveal a strong increase in the rate constants for all three displacement sequences. Specifically, this increase was more pronounced for the DS2 and the DS1 and somewhat smaller for the DS3. Structurally, deg is related to peg and provides a similar semi-hydrophobic environment without volume exclusion (i.e., no crowding effect) [
13]. For its structure and size, deg can be regarded as a co-solvent rather than a crowder, whose interactions with the nucleobases within the duplex (Bmut-CoBmut) facilitate the mismatched duplex displacement [
27]. These results confirm that hydrophobic interactions dominate crowding in enhancing displacement kinetics. The slight increase in the rate constant of displacement of the control sequence (DSCtrl) in deg buffer (
Figure 2c) shows that deg is interacting relatively strongly with the original duplex (ds), possibly changing the dipole-dipole interactions between the dyes and hence the FRET effect.
Since the different displacement strands showed distinct displacement kinetics in the different buffers and considering that the intracellular environment is a complex mixture of small molecules and macromolecules, [
28,
29] we measured the rate of the displacement kinetics in a mixed peg-deg buffer. Interestingly, for all three oligonucleotides, the rate of decay was faster compared with the peg-buffer and slower compared with the deg-buffer. These results suggest that there is a balance between solubility and crowding effects, with solubility exerting the dominant contribution.
Using the same experimental conditions, we investigated the strand displacement ability of all three oligonucleotides in a buffer containing pvp. We chose pvp because it is a structurally different macromolecule from peg and because of its polydisperse role as a crowder [
30]. Pvp is a biomaterial that is also used in tissue engineering and for the expansion of progenitor cells in vitro [
30,
31,
32]. From the time traces of strand displacement, it is evident that the rate of Cy5 fluorescence decay in pvp-buffer does not exceed the one in pure PBS. This observation agrees with previously reported results on macropolymers such as dextran, which lack the ability to increase the strand displacement constant of a DNA displacement strand. In addition, the displacement control sequence (DSCtrl) showed insignificant displacement kinetics in all different buffer conditions confirming the sequence specificity of the displacement process.
Previous reports showed that the duplex stability and melting temperature (
Tm) do not directly correlate with the rate of strand displacement by DNA strand in pure and peg-buffers. Specifically, increasing the NaCl concentration in the buffer compensates for the possible lower
Tm of the duplex caused by the peg crowder [
13]. Our data on the stabilized duplex (by addition of an extra 150 mM NaCl) did not show a change in the rank order of the strand displacement kinetics compared to the ones shown by DS1, DS2, and DS3 in the various buffers without additional salt. Moreover, the CD spectra confirm that the ds have overall B-DNA conformation in all crowder-enriched buffers, similar to pure PBS. The overall data indicate that even though the different crowders may react and intercalate within the duplex to affect the strand displacement ability, they do not directly destabilize and disturb the original duplex geometry.
As a final consideration, while we used a simplified single-step kinetic model to derive rate constants of displacement, the displacement process occurs in multiple steps, the first of which is diffusional. In conditions in which the diffusional step is rate limiting (e.g., low concentration of strands), the solvent viscosity can play an increasingly important role in the overall kinetics of the process. For this reason, we normalized the displacement kinetics by the solvent viscosity (
Table S1 and
Figure S4). As a result of this normalization, all buffers show increased rate constants of displacement compared to pure PBS, with the highest rate of displacement found in the peg-deg buffer. These results might suggest a cooperative effect between crowding and solubility, which is counteracted by the increase in solvent viscosity. Further investigations, a deeper mechanistic understanding of the displacement process, and a more comprehensive multi-step kinetic model will be required to verify this hypothesis.