Essential Role of Histidine for Rapid Copper(II)-Mediated Disassembly of Neurokinin B Amyloid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Estimating Aggregation Propensity
2.2. Thioflavin T Time-Resolved Fluorescence
2.3. Tryptophan Fluorescence
2.4. Electron Microscopy
2.5. Mass Spectroscopy (MS)
2.6. Modeling of a Two-Stranded β-Sheet
3. Results
3.1. An Aromatic Residue at the Third Position in NKB Is Required for Amyloid Formation
3.2. π–π Stacking Involving the Aromatic at the Third Position in NKB Contributes to Amyloid Formation
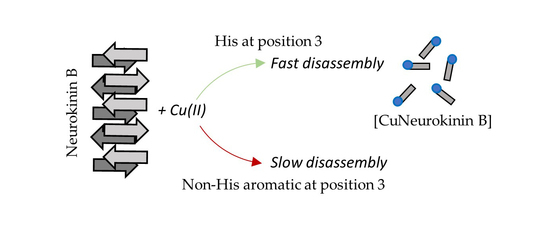
3.3. Histidine Is Required for Rapid Copper-Induced Disassembly
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Topaloglu, A.K.; Reimann, F.; Guclu, M.; Yalin, A.S.; Kotan, L.D.; Porter, K.M.; Serin, A.; Mungan, N.O.; Cook, J.R.; Ozbek, M.N.; et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 2009, 41, 354–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rance, N.E.; Krajewski, S.J.; Smith, M.A.; Cholanian, M.; Dacks, P.A. Neurokinin B and the hypothalamic regulation of reproduction. Brain Res. 2010, 1364, 116–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uenoyama, Y.; Nagae, M.; Tsuchida, H.; Inoue, N.; Tsukamura, H. Role of KNDy Neurons Expressing Kisspeptin, Neurokinin B, and Dynorphin A as a GnRH Pulse Generator Controlling Mammalian Reproduction. Front. Endocrinol. 2021, 12, 724632. [Google Scholar] [CrossRef]
- Anderson, R.A.; Millar, R.P. The roles of kisspeptin and neurokinin B in GnRH pulse generation in humans, and their potential clinical application. J. Neuroendocrinol. 2022, 34, e13081. [Google Scholar] [CrossRef] [PubMed]
- Rance, N.E.; Dacks, P.A.; Mittelman-Smith, M.A.; Romanovsky, A.A.; Krajewski-Hall, S.J. Modulation of body temperature and LH secretion by hypothalamic KNDy (kisspeptin, neurokinin B and dynorphin) neurons: A novel hypothesis on the mechanism of hot flushes. Front. Neuroendocrinol. 2013, 34, 211–227. [Google Scholar] [CrossRef] [Green Version]
- Jayasena, C.N.; Comninos, A.N.; Stefanopoulou, E.; Buckley, A.; Narayanaswamy, S.; Izzi-Engbeaya, C.; Abbara, A.; Ratnasabapathy, R.; Mogford, J.; Ng, N.; et al. Neurokinin B administration induces hot flushes in women. Sci. Rep. 2015, 5, 8466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modi, M.; Dhillo, W.S. Neurokinin 3 Receptor Antagonism: A Novel Treatment for Menopausal Hot Flushes. Neuroendocrinology 2019, 109, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Gasman, S.; Vitale, N. Lipid remodelling in neuroendocrine secretion. Biol. Cell 2017, 109, 381–390. [Google Scholar] [CrossRef]
- Persoon, C.M.; Moro, A.; Nassal, J.P.; Farina, M.; Broeke, J.H.; Arora, S.; Dominguez, N.; van Weering, J.R.; Toonen, R.F.; Verhage, M. Pool size estimations for dense-core vesicles in mammalian CNS neurons. EMBO J. 2018, 37, e99672. [Google Scholar] [CrossRef]
- Merighi, A. Costorage of High Molecular Weight Neurotransmitters in Large Dense Core Vesicles of Mammalian Neurons. Front. Cell. Neurosci. 2018, 12, 272. [Google Scholar] [CrossRef]
- Greenwald, J.; Riek, R. Biology of amyloid: Structure, function, and regulation. Structure 2010, 18, 1244–1260. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [Green Version]
- Chuang, E.; Hori, A.M.; Hesketh, C.D.; Shorter, J. Amyloid assembly and disassembly. J. Cell Sci. 2018, 131, jcs189928. [Google Scholar] [CrossRef] [Green Version]
- Balbach, J.J.; Ishii, Y.; Antzutkin, O.N.; Leapman, R.D.; Rizzo, N.W.; Dyda, F.; Reed, J.; Tycko, R. Amyloid fibril formation by A beta 16-22, a seven-residue fragment of the Alzheimer’s beta-amyloid peptide, and structural characterization by solid state NMR. Biochemistry 2000, 39, 13748–13759. [Google Scholar] [CrossRef] [PubMed]
- Azriel, R.; Gazit, E. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. An experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 2001, 276, 34156–34161. [Google Scholar] [CrossRef] [Green Version]
- Tenidis, K.; Waldner, M.; Bernhagen, J.; Fischle, W.; Bergmann, M.; Weber, M.; Merkle, M.L.; Voelter, W.; Brunner, H.; Kapurniotu, A. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 2000, 295, 1055–1071. [Google Scholar] [CrossRef] [Green Version]
- Makin, O.S.; Atkins, E.; Sikorski, P.; Johansson, J.; Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 2005, 102, 315–320. [Google Scholar] [CrossRef] [Green Version]
- Tatko, C.D.; Waters, M.L. Selective aromatic interactions in beta-hairpin peptides. J. Am. Chem. Soc. 2002, 124, 9372–9373. [Google Scholar] [CrossRef]
- Gazit, E. Self assembly of short aromatic peptides into amyloid fibrils and related nanostructures. Prion 2007, 1, 32–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayawardena, B.M.; Jones, M.R.; Hong, Y.; Jones, C.E. Copper ions trigger disassembly of neurokinin B functional amyloid and inhibit de novo assembly. J. Struct. Biol. 2019, 208, 107394. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xia, N.; Chen, M.; Yang, P.; Liu, L.; Wang, J. A copper complex formed with neurokinin B: Binding stoichiometry, redox properties, self-assembly and cytotoxicity. Metallomics 2020, 12, 1802–1810. [Google Scholar] [CrossRef] [PubMed]
- Krishna Deepak, R.N.; Sankararamakrishnan, R. N-H⋯N Hydrogen Bonds Involving Histidine Imidazole Nitrogen Atoms: A New Structural Role for Histidine Residues in Proteins. Biochemistry 2016, 55, 3774–3783. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Du, Q.S.; Meng, J.Z.; Pang, Z.W.; Huang, R.B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Harford, C.; Sarkar, B. Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: Metal binding, DNA cleavage, and other properties. Acc. Chem. Res. 1997, 30, 123–130. [Google Scholar] [CrossRef]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Mold, M.; Ouro-Gnao, L.; Wieckowski, B.M.; Exley, C. Copper prevents amyloid-beta(1-42) from forming amyloid fibrils under near-physiological conditions in vitro. Sci. Rep. 2013, 3, 1256. [Google Scholar] [CrossRef] [Green Version]
- House, E.; Mold, M.; Collingwood, J.; Baldwin, A.; Goodwin, S.; Exley, C. Copper abolishes the beta-sheet secondary structure of preformed amyloid fibrils of amyloid-beta(42). J. Alzheimers Dis. 2009, 18, 811–817. [Google Scholar] [CrossRef] [Green Version]
- Senko, M.W. IsoPro Computer Program 3.0. Available online: https://sites.google.com/site/isoproms/ (accessed on 21 October 2022).
- Biancalana, M.; Makabe, K.; Koide, A.; Koide, S. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J. Mol. Biol. 2009, 385, 1052–1063. [Google Scholar] [CrossRef]
- Vivian, J.T.; Callis, P.R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001, 80, 2093–2109. [Google Scholar] [CrossRef] [Green Version]
- Wachter, R.M.; Elsliger, M.A.; Kallio, K.; Hanson, G.T.; Remington, S.J. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure 1998, 6, 1267–1277. [Google Scholar] [CrossRef] [Green Version]
- Touchette, J.C.; Williams, L.L.; Ajit, D.; Gallazzi, F.; Nichols, M.R. Probing the amyloid-beta(1-40) fibril environment with substituted tryptophan residues. Arch. Biochem. Biophys. 2010, 494, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R. Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron 2015, 86, 632–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iadanza, M.G.; Silvers, R.; Boardman, J.; Smith, H.I.; Karamanos, T.K.; Debelouchina, G.T.; Su, Y.; Griffin, R.G.; Ranson, N.A.; Radford, S.E. The structure of a beta2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Commun. 2018, 9, 4517. [Google Scholar] [CrossRef] [Green Version]
- Seuring, C.; Verasdonck, J.; Ringler, P.; Cadalbert, R.; Stahlberg, H.; Bockmann, A.; Meier, B.H.; Riek, R. Amyloid Fibril Polymorphism: Almost Identical on the Atomic Level, Mesoscopically Very Different. J. Phys. Chem. B 2017, 121, 1783–1792. [Google Scholar] [CrossRef]
- Stokowa-Soltys, K.; Szczerba, K.; Pacewicz, M.; Wieczorek, R.; Wezynfeld, N.E.; Bal, W. Interactions of neurokinin B with copper(II) ions and their potential biological consequences. Dalton Trans. 2022, 51, 14267–14276. [Google Scholar] [CrossRef]
- Russino, D.; McDonald, E.; Hejazi, L.; Hanson, G.R.; Jones, C.E. The Tachykinin Peptide Neurokinin B Binds Copper Forming an Unusual [CuII(NKB)] Complex and Inhibits Copper Uptake into 1321N1 Astrocytoma Cells. ACS Chem. Neurosci. 2013, 4, 1371–1381. [Google Scholar] [CrossRef] [Green Version]
- Peacey, L.; Elphick, M.R.; Jones, C.E. Roles of copper in neurokinin B and gonadotropin-releasing hormone structure and function and the endocrinology of reproduction. Gen. Comp. Endocrinol. 2019, 287, 113342. [Google Scholar] [CrossRef]
- Bryce, G.F.; Gurd, F.R. Visible spectra and optical rotatory properties of cupric ion complexes of L-histidine-containing peptides. J. Biol. Chem. 1966, 241, 122–129. [Google Scholar] [CrossRef]
- Rozga, M.; Protas, A.M.; Jablonowska, A.; Dadlez, M.; Bal, W. The Cu(II) complex of Abeta40 peptide in ammonium acetate solutions. Evidence for ternary species formation. Chem. Commun. 2009, 2009, 1374–1376. [Google Scholar] [CrossRef] [PubMed]
- Nagaj, J.; Stokowa-Soltys, K.; Kurowska, E.; Fraczyk, T.; Jezowska-Bojczuk, M.; Bal, W. Revised coordination model and stability constants of Cu(II) complexes of tris buffer. Inorg. Chem. 2013, 52, 13927–13933. [Google Scholar] [CrossRef] [PubMed]
- Nespovitaya, N.; Gath, J.; Barylyuk, K.; Seuring, C.; Meier, B.H.; Riek, R. Dynamic Assembly and Disassembly of Functional beta-Endorphin Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 846–856. [Google Scholar] [CrossRef]
- Flashner, E.; Raviv, U.; Friedler, A. The effect of tachykinin neuropeptides on amyloid beta aggregation. Biochem. Biophys. Res. Commun. 2011, 407, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Yankner, B.A.; Duffy, L.K.; Kirschner, D.A. Neurotrophic and neurotoxic effects of amyloid beta protein: Reversal by tachykinin neuropeptides. Science 1990, 250, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.A.; Singh, J.; Thornton, J.M. Pi-pi interactions: The geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J. Mol. Biol. 1991, 218, 837–846. [Google Scholar] [CrossRef]
- Stankovic, I.M.; Bozinovski, D.M.; Brothers, E.N.; Belic, M.R.; Hall, M.B.; Zaric, S.D. Interactions of Aromatic Residues in Amyloids: A Survey of Protein Data Bank Crystallographic Data. Cryst. Growth Des. 2017, 17, 6353–6362. [Google Scholar] [CrossRef]
- Bossak-Ahmad, K.; Wisniewska, M.D.; Bal, W.; Drew, S.C.; Fraczyk, T. Ternary Cu(II) Complex with GHK Peptide and Cis-Urocanic Acid as a Potential Physiologically Functional Copper Chelate. Int. J. Mol. Sci. 2020, 21, 6190. [Google Scholar] [CrossRef]
- Valery, C.; Deville-Foillard, S.; Lefebvre, C.; Taberner, N.; Legrand, P.; Meneau, F.; Meriadec, C.; Delvaux, C.; Bizien, T.; Kasotakis, E.; et al. Atomic view of the histidine environment stabilizing higher-pH conformations of pH-dependent proteins. Nat. Commun. 2015, 6, 7771. [Google Scholar] [CrossRef]




| Name | Sequence | HydroP 1 | %Agg 2 |
|---|---|---|---|
| NKB | DMHDFFVGLM-NH2 | 0.68 | 100 |
| H3T | DMTDFFVGLM-NH2 | 0.93 | 93.2 |
| H3A | DMADFFVGLM-NH2 | 1.18 | 93.1 |
| H3W | DMWDFFVGLM-NH2 | 0.91 | 93.3 |
| H3F | DMFDFFVGLM-NH2 | 1.28 | 93.4 |
| F5W | DMHDWFVGLM-NH2 | 0.31 | 59.5 |
| F5,6W | DMHDWWVGLM-NH2 | 0.30 | 26.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayawardena, B.M.; Peacey, L.; Gamsjaeger, R.; Jones, C.E. Essential Role of Histidine for Rapid Copper(II)-Mediated Disassembly of Neurokinin B Amyloid. Biomolecules 2022, 12, 1585. https://doi.org/10.3390/biom12111585
Jayawardena BM, Peacey L, Gamsjaeger R, Jones CE. Essential Role of Histidine for Rapid Copper(II)-Mediated Disassembly of Neurokinin B Amyloid. Biomolecules. 2022; 12(11):1585. https://doi.org/10.3390/biom12111585
Chicago/Turabian StyleJayawardena, Bhawantha M., Lorraine Peacey, Roland Gamsjaeger, and Christopher E. Jones. 2022. "Essential Role of Histidine for Rapid Copper(II)-Mediated Disassembly of Neurokinin B Amyloid" Biomolecules 12, no. 11: 1585. https://doi.org/10.3390/biom12111585