Saccharomyces cerevisiae as a Toolkit for COP9 Signalosome Research
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Plasmid Cloning
2.3. Growth Conditions
2.4. Native Protein Extraction
2.5. Calmodulin-Based Affinity Purification of the CSN
2.6. Immunoblotting
2.7. Purification of the Substrate by Ni-NTA Affinity Chromatography
2.8. Fluorescence Microscopy
2.9. Ergosterol Extraction and Analysis
2.10. Endogenous Oxidative Stress Measurements
2.11. CSN Activity Assay
3. Results and Discussion
3.1. S. cerevisiae Harbors a Highly Conserved Enzymatic Core Complex
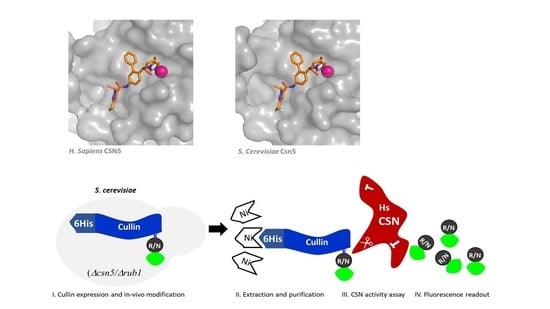
3.2. Utilizing the S. cerevisiae Neddylation Pathway to Develop a Fluorogenic CSN Substrate for In Vitro Studies
3.3. CSN5i-3 Targets CSN5/Csn5 from Diverged Eukaryotic Sources
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Adapting proteostasis for disease intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef]
- Deshaies, R.J. Structural biology: Corralling a protein-degradation regulator. Nature 2014, 512, 145–146. [Google Scholar] [CrossRef]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef]
- Reinstein, E.; Ciechanover, A. Narrative review: Protein degradation and human diseases: The ubiquitin connection. Ann. Intern. Med. 2006, 145, 676–684. [Google Scholar] [CrossRef]
- Nedelsky, N.B.; Todd, P.K.; Taylor, J.P. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim. Biophys. Acta 2008, 1782, 691–699. [Google Scholar] [CrossRef]
- Ciechanover, A. Intracellular protein degradation: From a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorg. Med. Chem. 2013, 21, 3400–3410. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Gelmann, E.P. The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 2005, 23, 4776–4789. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2017. [Google Scholar] [CrossRef]
- Hu, H.Y. Protein Ubiquitination and Deubiquitination. Curr. Protein Pept. Sci. 2012, 13, 413. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Suzuki, T.; Chiba, T. The ligation systems for ubiquitin and ubiquitin-like proteins. Mol. Cells 1998, 8, 503–512. [Google Scholar]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The Ubiquitin-Proteasome System of Saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Finley, D. A proteasome for all occasions. FEBS Lett. 2007, 581, 2854–2861. [Google Scholar] [CrossRef]
- Pick, E.; Hofmann, K.; Glickman, M.H. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell 2009, 35, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Scheel, H.; Hofmann, K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinform. 2005, 6, 71. [Google Scholar] [CrossRef]
- Ambroggio, X.I.; Rees, D.C.; Deshaies, R.J. JAMM: A Metalloprotease-Like Zinc Site in the Proteasome and Signalosome. PLoS Biology 2004, 2, e2. [Google Scholar] [CrossRef]
- Schmaler, T.; Dubiel, W. Control of Deneddylation by the COP9 Signalosome. Subcell. Biochem. 2010, 54, 57–68. [Google Scholar] [CrossRef]
- Maytal-Kivity, V.; Reis, N.; Hofmann, K.; Glickman, M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002, 3, 28. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., III; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 Metalloprotease in Deubiquitination and Degradation by the 26S Proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of Predicted Metalloprotease Motif of Jab1/Csn5 in Cleavage of NEDD8 from CUL1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.; Wolf, D.A.; Wei, N.; Shevchenko, A.; Deshaies, R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001, 292, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Krist, D.T.; Prabu, J.R.; Hill, S.; Klugel, M.; Neumaier, L.M.; von Gronau, S.; Kleiger, G.; Schulman, B.A. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 2020, 578, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Toth, J.I.; Yang, L.; Dahl, R.; Petroski, M.D. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 2012, 1, 309–316. [Google Scholar] [CrossRef]
- Schlierf, A.; Altmann, E.; Quancard, J.; Jefferson, A.B.; Assenberg, R.; Renatus, M.; Jones, M.; Hassiepen, U.; Schaefer, M.; Kiffe, M.; et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat. Commun. 2016, 7, 13166. [Google Scholar] [CrossRef]
- Wei, N.; Chamovitz, D.A.; Deng, X.W. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of plant development. Cell 1994, 78, 117–124. [Google Scholar] [CrossRef]
- Chamovitz, D.A.; Wei, N.; Osterlund, M.T.; von Arnim, A.G.; Staub, J.M.; Matsui, M.; Deng, X.W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant development switch. Cell 1996, 86, 115–121. [Google Scholar] [CrossRef]
- Seeger, M.; Gordon, C.; Ferrell, K.; Dubiel, W. Characteristics of 26S proteases from fission yeast mutants which arrest in mitosis. J. Mol. Biol. 1996, 263, 423–431. [Google Scholar] [CrossRef]
- Wei, N.; Serino, G.; Deng, X.W. The COP9 signalosome: More than a protease. Trends Biochem. Sci. 2008, 33, 592–600. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Isono, E. The COP9 signalosome and its role in plant development. Eur. J. Cell Biol. 2010, 89, 157–162. [Google Scholar] [CrossRef]
- Oron, E.; Mannervik, M.; Rencus, S.; Harari-Steinberg, O.; Neuman-Silberberg, S.; Segal, D.; Chamovitz, D.A. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 2002, 129, 4399–4409. [Google Scholar]
- Zhao, R.; Yeung, S.C.; Chen, J.; Iwakuma, T.; Su, C.H.; Chen, B.; Qu, C.; Zhang, F.; Chen, Y.T.; Lin, Y.L.; et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J. Clin. Investig. 2011, 121, 851–865. [Google Scholar] [CrossRef]
- Braus, G.H.; Irniger, S.; Bayram, O. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 2010, 13, 672–676. [Google Scholar] [CrossRef]
- Pick, E. The necessity of NEDD8/Rub1 for vitality and its association with mitochondria-derived oxidative stress. Redox Biol. 2020, 37, 101765. [Google Scholar] [CrossRef] [PubMed]
- Cavadini, S.; Fischer, E.S.; Bunker, R.D.; Potenza, A.; Lingaraju, G.M.; Goldie, K.N.; Mohamed, W.I.; Faty, M.; Petzold, G.; Beckwith, R.E.; et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 2016, 531, 598–603. [Google Scholar] [CrossRef]
- Lingaraju, G.M.; Bunker, R.D.; Cavadini, S.; Hess, D.; Hassiepen, U.; Renatus, M.; Fischer, E.S.; Thoma, N.H. Crystal structure of the human COP9 signalosome. Nature 2014, 512, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Mosadeghi, R.; Reichermeier, K.M.; Winkler, M.; Schreiber, A.; Reitsma, J.M.; Zhang, Y.; Stengel, F.; Cao, J.; Kim, M.; Sweredoski, M.J.; et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.Y.; Yoneda-Kato, N. Mammalian COP9 signalosome. Genes Cells 2009, 14, 1209–1225. [Google Scholar] [CrossRef]
- Liu, C.; Guo, L.Q.; Menon, S.; Jin, D.; Pick, E.; Wang, X.; Deng, X.W.; Wei, N. COP9 signalosome subunit Csn8 is involved in maintaining proper duration of the G1 phase. J. Biol. Chem. 2013, 288, 20443–20452. [Google Scholar] [CrossRef]
- Claret, F.X.; Hibi, M.; Dhut, S.; Toda, T.; Karin, M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 1996, 383, 453–457. [Google Scholar] [CrossRef]
- Yoshida, A.; Yoneda-Kato, N.; Kato, J.Y. CSN5 specifically interacts with CDK2 and controls senescence in a cytoplasmic cyclin E-mediated manner. Sci. Rep. 2013, 3, 1054. [Google Scholar] [CrossRef]
- Tomoda, K.; Kato, J.Y.; Tatsumi, E.; Takahashi, T.; Matsuo, Y.; Yoneda-Kato, N. The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood 2005, 105, 775–783. [Google Scholar] [CrossRef]
- Pick, E.; Golan, A.; Zimbler, J.Z.; Guo, L.; Sharaby, Y.; Tsuge, T.; Hofmann, K.; Wei, N. The Minimal Deneddylase Core of the COP9 Signalosome Excludes the Csn6 MPN(-) Domain. PLoS ONE 2012, 7, e43980. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Kleifeld, O.; Lande-Atir, A.; Bsoul, M.; Kleiman, M.; Krutauz, D.; Book, A.; Vierstra, R.D.; Hofmann, K.; Reis, N.; et al. Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell 2011, 22, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Maytal-Kivity, V.; Pick, E.; Piran, R.; Hofmann, K.; Glickman, M.H. The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int. J. Biochem. Cell Biol. 2003, 35, 706–715. [Google Scholar] [CrossRef]
- Maytal-Kivity, V.; Piran, R.; Pick, E.; Hofmann, K.; Glickman, M.H. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 2002, 12, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Israeli, R.; Cirigliano, A.; Gihaz, S.; Trabelcy, B.; Braus, G.H.; Gerchman, Y.; Fishman, A.; Negri, R.; Rinaldi, T.; et al. The COP9 signalosome mediates the Spt23 regulated fatty acid desaturation and ergosterol biosynthesis. FASEB J. 2020. [Google Scholar] [CrossRef]
- Bramasole, L.; Sinha, A.; Gurevich, S.; Radzinski, M.; Klein, Y.; Panat, N.; Gefen, E.; Rinaldi, T.; Jimenez-Morales, D.; Johnson, J.; et al. Proteasome lid bridges mitochondrial stress with Cdc53/Cullin1 NEDDylation status. Redox Biol. 2019, 20, 533–543. [Google Scholar] [CrossRef]
- Liakopoulos, D.; Doenges, G.; Matuschewski, K.; Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998, 17, 2208–2214. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, D.; Busgen, T.; Brychzy, A.; Jentsch, S.; Pause, A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl. Acad. Sci. USA 1999, 96, 5510–5515. [Google Scholar] [CrossRef]
- Eisele, M.R.; Reed, R.G.; Rudack, T.; Schweitzer, A.; Beck, F.; Nagy, I.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Tomko, R.J., Jr.; et al. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep. 2018, 24, 1301–1315.e1315. [Google Scholar] [CrossRef]
- Wehmer, M.; Rudack, T.; Beck, F.; Aufderheide, A.; Pfeifer, G.; Plitzko, J.M.; Forster, F.; Schulten, K.; Baumeister, W.; Sakata, E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2017, 114, 1305–1310. [Google Scholar] [CrossRef]
- Dessau, M.; Halimi, Y.; Erez, T.; Chomsky-Hecht, O.; Chamovitz, D.A.; Hirsch, J.A. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell 2008, 20, 2815–2834. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.; Hetfeld, B.; Dubiel, W.; Wolf, D.A. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002, 3, 15. [Google Scholar] [CrossRef]
- Unverdorben, P.; Beck, F.; Śledź, P.; Schweitzer, A.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Förster, F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, 5544–5549. [Google Scholar] [CrossRef]
- Faull, S.V.; Lau, A.M.C.; Martens, C.; Ahdash, Z.; Hansen, K.; Yebenes, H.; Schmidt, C.; Beuron, F.; Cronin, N.B.; Morris, E.P.; et al. Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat. Commun. 2019, 10, 3814. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Benkert, P.; Tosatto, S.C.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins 2008, 71, 261–277. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Michnick, S.W.; Ear, P.H.; Landry, C.; Malleshaiah, M.K.; Messier, V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010, 470, 335–368. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.Q. The cullin protein family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, I.W.; Rabut, G.; Poveda, A.; Scheel, H.; Malmstrom, J.; Ulrich, H.; Hofmann, K.; Pasero, P.; Peter, M.; Luke, B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008, 9, 1034–1040. [Google Scholar] [CrossRef]
- Patton, E.E.; Willems, A.R.; Sa, D.; Kuras, L.; Thomas, D.; Craig, K.L.; Tyers, M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998, 12, 692–705. [Google Scholar] [CrossRef]
- Rabut, G.; Le Dez, G.; Verma, R.; Makhnevych, T.; Knebel, A.; Kurz, T.; Boone, C.; Deshaies, R.J.; Peter, M. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol. Cell 2011, 43, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.C.; Monda, J.K.; Grace, C.R.; Duda, D.M.; Kriwacki, R.W.; Kurz, T.; Schulman, B.A. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell 2010, 39, 784–796. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 1970, 66, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Kurz, T.; Ozlu, N.; Rudolf, F.; O’Rourke, S.M.; Luke, B.; Hofmann, K.; Hyman, A.A.; Bowerman, B.; Peter, M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 2005, 435, 1257–1261. [Google Scholar] [CrossRef]
- Cotto-Rios, X.M.; Bekes, M.; Chapman, J.; Ueberheide, B.; Huang, T.T. Deubiquitinases as a signaling target of oxidative stress. Cell Rep. 2012, 2, 1475–1484. [Google Scholar] [CrossRef]
- Leidecker, O.; Matic, I.; Mahata, B.; Pion, E.; Xirodimas, D.P. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 2012, 11, 1142–1150. [Google Scholar] [CrossRef]
- Keuss, M.J.; Hjerpe, R.; Hsia, O.; Gourlay, R.; Burchmore, R.; Trost, M.; Kurz, T. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 2019, 38, e100024. [Google Scholar] [CrossRef] [PubMed]
- Golan, A.; Wei, N.; Pick, E. Immunodepletion and Immunopurification as Approaches for CSN Research. Methods Mol. Biol. 2016, 1449, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Cirigliano, A.; Stirpe, A.; Menta, S.; Mori, M.; Dell’Edera, D.; Pick, E.; Negri, R.; Botta, B.; Rinaldi, T. Yeast as a tool to select inhibitors of the cullin deneddylating enzyme Csn5. J. Enzyme Inhib. Med. Chem. 2016, 31, 1632–1637. [Google Scholar] [CrossRef] [PubMed]






| H. sapiens vs. S. cerevisiae | CSN1 vs. Csn11 [*1] | CSN4 vs. Rpn5 [*2] | CSN5 vs. Csn5 [*3] | CSN6 vs. Csi1 [*4] | CSN7 vs. Csn9 [*5] | CSN2 vs. Csn10 [*6] |
|---|---|---|---|---|---|---|
| Sequence identity (%) | 16% | 100% to the 19S lid Rpn5 | 39.3% | 15% | 23.4% | 20% |
| Coverage (%) | 85% | 100% | 47% | 82% | 77% | 84% |
| GMQE | 0.38 | 0.76 | 0.25 | 0.5 | 0.39 | 0.12 |
| QMEAN | −5.57 | −2.61 | −0.62 | −4.08 | −1.2 | −4.7 |
| ** Confidence (%) | 99.7% | 98.0% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harshuk-Shabso, D.; Castel, N.; Israeli, R.; Harari, S.; Pick, E. Saccharomyces cerevisiae as a Toolkit for COP9 Signalosome Research. Biomolecules 2021, 11, 497. https://doi.org/10.3390/biom11040497
Harshuk-Shabso D, Castel N, Israeli R, Harari S, Pick E. Saccharomyces cerevisiae as a Toolkit for COP9 Signalosome Research. Biomolecules. 2021; 11(4):497. https://doi.org/10.3390/biom11040497
Chicago/Turabian StyleHarshuk-Shabso, Dana, Noam Castel, Ran Israeli, Sheri Harari, and Elah Pick. 2021. "Saccharomyces cerevisiae as a Toolkit for COP9 Signalosome Research" Biomolecules 11, no. 4: 497. https://doi.org/10.3390/biom11040497