Digital Pathology Enables Automated and Quantitative Assessment of Inflammatory Activity in Patients with Chronic Liver Disease
Abstract
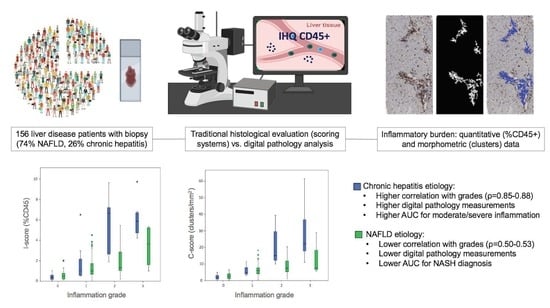
:1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Histological Analysis
2.3. Digital Image Analysis
2.4. Statistical Analysis
3. Results
3.1. Patients and Histological Characteristics
3.2. Digital Image Analysis and Inflammation Scoring Systems
3.3. Relationship between Different Histological Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1


Appendix A.2

Appendix A.3
| Etiology | Histologic Grades | QuPath % Cut-Off | QuPath AUC (95%CI) | DeLong p-Value vs. %CD45 | DeLong p-Value vs. Clusters |
|---|---|---|---|---|---|
| Grade ≥ 1 | >5.0 | 0.62 (0.52–0.71) | <0.001 | <0.001 | |
| NAFLD | Grade ≥ 2 | >7.4 | 0.67 (0.58–0.77) | 0.007 | 0.02 |
| Grade ≥ 3 | >15.1 | 0.85 (0.70–0.99) | 0.59 | 0.98 | |
| Grade ≥ 1 | >5.5 | 0.63 (0.53–0.72) | 0.003 | 0.005 | |
| CH | Grade ≥ 2 | >7.3 | 0.78 (0.70–0.86) | 0.06 | 0.02 |
| Grade ≥ 3 | >11.1 | 0.87 (0.77–0.96) | 0.47 | 0.06 |

References
- GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef] [Green Version]
- Davison, B.A.; Harrison, S.A.; Cotter, G.; Alkhouri, N.; Sanyal, A.; Edwards, C.; Colca, J.R.; Iwashita, J.; Koch, G.G.; Dittrich, H.C. Suboptimal reliability of liver biopsy evaluation has implications for randomized clinical trials. J. Hepatol. 2020, 73, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Standish, R.; Cholongitas, E.; Dhillon, A.; Burroughs, A.K.; Dhillon, A.P. An appraisal of the histopathological assessment of liver fibrosis. Gut 2006, 55, 569–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishak, K.; Baptista, A.; Bianchi, L.; Callea, F.; De Groote, J.; Gudat, F.; Denk, H.; Desmet, V.; Korb, G.; Macsween, R.N.; et al. Histological grading and staging of chronic hepatitis. J. Hepatol. 1995, 22, 696–699. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Stepanova, M.; Rafiq, N.; Henry, L.; Loomba, R.; Makhlouf, H.; Goodman, Z. Nonalcoholic steatofibrosis independently predicts mortality in nonalcoholic fatty liver disease. Hepatol. Commun. 2017, 1, 421–428. [Google Scholar] [CrossRef]
- Loomba, R.; Chalasani, N. The Hierarchical Model of NAFLD: Prognostic Significance of Histologic Features in NASH. Gastroenterology 2015, 149, 278–281. [Google Scholar] [CrossRef]
- Goodman, Z.D. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J. Hepatol. 2007, 47, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Melo, R.C.N.; Raas, M.W.D.; Palazzi, C.; Neves, V.H.; Malta, K.K.; Silva, T.P. Whole Slide Imaging and Its Applications to Histopathological Studies of Liver Disorders. Front. Med. 2020, 6, 310. [Google Scholar] [CrossRef] [Green Version]
- Paradis, V.; Quaglia, A. Digital pathology, what is the future? J. Hepatol. 2019, 70, 1016–1018. [Google Scholar] [CrossRef] [Green Version]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Forlano, R.; Mullish, B.H.; Giannakeas, N.; Maurice, J.B.; Angkathunyakul, N.; Lloyd, J.; Tzallas, A.T.; Tsipouras, M.; Yee, M.; Thursz, M.R.; et al. High-Throughput, Machine Learning–Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2020, 18, 2081–2090.e9. [Google Scholar] [CrossRef] [Green Version]
- Taylor-Weiner, A.; Pokkalla, H.; Han, L.; Jia, C.; Huss, R.; Chung, C.; Elliott, H.; Glass, B.; Pethia, K.; Carrasco-Zevallos, O.; et al. A Machine Learning Approach Enables Quantitative Measurement of Liver Histology and Disease Monitoring in NASH. Hepatology 2021, 74, 133–147. [Google Scholar] [CrossRef]
- Vanderbeck, S.; Bockhorst, J.; Kleiner, D.; Komorowski, R.; Chalasani, N.; Gawrieh, S. Automatic quantification of lobular inflammation and hepatocyte ballooning in nonalcoholic fatty liver disease liver biopsies. Hum. Pathol. 2015, 46, 767–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stasi, C.; Tsochatzis, E.A.; Hall, A.; Rosenberg, W.; Milani, S.; Dhillon, A.P.; Pinzani, M. Comparison and correlation of fibrosis stage assessment by collagen proportionate area (CPA) and the ELF panel in patients with chronic liver disease. Dig. Liver Dis. 2019, 51, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Buzzetti, E.; Hall, A.; Ekstedt, M.; Manuguerra, R.; Misas, M.G.; Covelli, C.; Leandro, G.; Luong, T.; Kechagias, S.; Manesis, E.K.; et al. Collagen proportionate area is an independent predictor of long-term outcome in patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2019, 49, 1214–1222. [Google Scholar] [CrossRef]
- Masugi, Y.; Abe, T.; Tsujikawa, H.; Effendi, K.; Hashiguchi, A.; Abe, M.; Imai, Y.; Hino, K.; Hige, S.; Kawanaka, M.; et al. Quantitative assessment of liver fibrosis reveals a nonlinear association with fibrosis stage in nonalcoholic fatty liver disease. Hepatol. Commun. 2017, 2, 58–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marti-Aguado, D.; Rodríguez-Ortega, A.; Mestre-Alagarda, C.; Bauza, M.; Valero-Pérez, E.; Alfaro-Cervello, C.; Benlloch, S.; Pérez-Rojas, J.; Ferrández, A.; Alemany-Monraval, P.; et al. Digital pathology: Accurate technique for quantitative assessment of histological features in metabolic-associated fatty liver disease. Aliment. Pharmacol. Ther. 2020, 53, 160–171. [Google Scholar] [CrossRef]
- E Torlakovic, E.; Naresh, K.; Kremer, M.; Porwit, A.; Van Der Walt, J.; Hyjek, E. Call for a European programme in external quality assurance for bone marrow immunohistochemistry; report of a European Bone Marrow Working Group pilot study. J. Clin. Pathol. 2009, 62, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.-C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Bedossa, P.; Poynard, T. The French METAVIR Cooperative Study Group. An algorithm for grading activity in chronic hepatitis C. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Wilson, L.A.; Behling, C.; Guy, C.; Contos, M.; Cummings, O.; Yeh, M.; Gill, R.; Chalasani, N.; et al. Association of Histologic Disease Activity With Progression of Nonalcoholic Fatty Liver Disease. JAMA Netw. Open 2019, 2, e1912565. [Google Scholar] [CrossRef] [Green Version]
- Ghany, M.G.; Kleiner, D.; Alter, H.; Doo, E.; Khokar, F.; Promrat, K.; Herion, D.; Park, Y.; Liang, T.; Hoofnagle, J.H. Progression of fibrosis in chronic hepatitis C. Gastroenterology 2003, 124, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Eng. Med. Boil. Mag. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Tsochatzis, E.; Bruno, S.; Isgro, G.; Hall, A.; Theocharidou, E.; Manousou, P.; Dhillon, A.P.; Burroughs, A.K.; Luong, T.V. Collagen proportionate area is superior to other histological methods for sub-classifying cirrhosis and determining prognosis. J. Hepatol. 2014, 60, 948–954. [Google Scholar] [CrossRef] [Green Version]
- Calvaruso, V.; Burroughs, A.K.; Standish, R.; Manousou, P.; Grillo, F.; Leandro, G.; Maimone, S.; Pleguezuelo, M.; Xirouchakis, I.; Guerrini, G.P.; et al. Computer-assisted image analysis of liver collagen: Relationship to Ishak scoring and hepatic venous pressure gradient. Hepatology 2009, 49, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Israelsen, M.; Misas, M.G.; Koutsoumourakis, A.; Huang, Y.; Thiele, M.; Hall, A.; Rasmussen, D.; Covelli, C.; Buzzetti, E.; Prat, L.I.; et al. Collagen proportionate area predicts clinical outcomes in patients with alcohol-related liver disease. Aliment. Pharmacol. Ther. 2020, 52, 1728–1739. [Google Scholar]
- Nascimbeni, F.; Ballestri, S.; Machado, M.V.; Mantovani, A.; Cortez-Pinto, H.; Targher, G.; Lonardo, A. Clinical relevance of liver histopathology and different histological classifications of NASH in adults. Expert Rev. Gastroenterol. Hepatol. 2017, 12, 351–367. [Google Scholar] [CrossRef]
- Bianconi, F.; Kather, J.N.; Reyes-Aldasoro, C.C. Experimental Assessment of Color Deconvolution and Color Normalization for Automated Classification of Histology Images Stained with Hematoxylin and Eosin. Cancers 2020, 12, 3337. [Google Scholar] [CrossRef]
- Roy, M.; Wang, F.; Vo, H.; Teng, D.; Teodoro, G.; Farris, A.B.; Castillo-Leon, E.; Vos, M.B.; Kong, J. Deep-learning-based accurate hepatic steatosis quantification for histological assessment of liver biopsies. Lab. Investig. 2020, 100, 1367–1383. [Google Scholar] [CrossRef]
- Hall, A.; Germani, G.; Isgrò, G.; Burroughs, A.K.; Dhillon, A.P. Fibrosis distribution in explanted cirrhotic livers. Histopathology 2011, 60, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Rosselli, M.; Macnaughtan, J.; Jalan, R.; Pinzani, M. Beyond scoring: A modern interpretation of disease progression in chronic liver disease. Gut 2013, 62, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Long, M.T.; Gandhi, S.; Loomba, R. Advances in non-invasive biomarkers for the diagnosis and monitoring of non-alcoholic fatty liver disease. Metabolism 2020, 111, 154259. [Google Scholar] [CrossRef] [PubMed]
- Newsome, P.N.; Sasso, M.; Deeks, J.J.; Paredes, A.; Boursier, J.; Chan, W.K.; Yilmaz, Y.; Czernichow, S.; Zheng, M.H.; Wong, V.W.S.; et al. FibroScan-AST (FAST) score for the non-invasive identification of patients with non-alcoholic steatohepatitis with significant activity and fibrosis: A prospective derivation and global validation study. Lancet Gastroenterol. Hepatol. 2020, 5, 362–373, Correction in Lancet Gastroenterol. Hepatol. 2020, 5, e3. [Google Scholar] [CrossRef] [Green Version]
- Schwenger, K.J.; Chen, L.; Chelliah, A.; Da Silva, H.E.; Teterina, A.; Comelli, E.M.; Taibi, A.; Arendt, B.M.; Fischer, S.; Allard, J.P. Markers of activated inflammatory cells are associated with disease severity and intestinal microbiota in adults with non-alcoholic fatty liver disease. Int. J. Mol. Med. 2018, 42, 2229–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ampuero, J.; Aller, R.; Gallego-Durán, R.; Crespo, J.; Abad, J.; González-Rodríguez, Á.; Gómez-Camarero, J.; Caballería, J.; Iacono, O.L.; Ibañez, L.; et al. Definite and indeterminate nonalcoholic steatohepatitis share similar clinical features and prognosis: A longitudinal study of 1893 biopsy-proven nonalcoholic fatty liver disease subjects. Liver Int. 2021, 41, 2076–2086. [Google Scholar] [CrossRef]
- Pelusi, S.; Cespiati, A.; Rametta, R.; Pennisi, G.; Mannisto, V.; Rosso, C.; Baselli, G.A.; Dongiovanni, P.; Fracanzani, A.L.; Badiali, S.; et al. Prevalence and Risk Factors of Significant Fibrosis in Patients With Nonalcoholic Fatty Liver Without Steatohepatitis. Clin. Gastroenterol. Hepatol. 2019, 17, 2310–2319.e6. [Google Scholar] [CrossRef] [Green Version]
- Tacke, F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017, 66, 1300–1312. [Google Scholar] [CrossRef]
- Brunt, E.M.; Kleiner, D.E.; Wilson, L.A.; Unalp, A.; Behling, C.E.; Lavine, J.E.; Neuschwander-Tetri, B.A. NASH Clinical Research Network Portal chronic inflammation in nonalcoholic fatty liver disease (NAFLD): A histologic marker of advanced NAFLD-Clinicopathologic correlations from the nonalcoholic steatohepatitis clinical research network. Hepatology 2009, 49, 809–820. [Google Scholar] [CrossRef] [Green Version]
- Bowlus, C.L.; Pockros, P.J.; Kremer, A.E.; Parés, A.; Forman, L.M.; Drenth, J.P.; Ryder, S.D.; Terracciano, L.; Jin, Y.; Liberman, A.; et al. Long-Term Obeticholic Acid Therapy Improves Histological Endpoints in Patients With Primary Biliary Cholangitis. Clin. Gastroenterol. Hepatol. 2020, 18, 1170–1178.e6. [Google Scholar] [CrossRef]




| Characteristic | Overall Cohort (n = 156) |
|---|---|
| Female sex | 91 (58%) |
| BMI (kg/m2) Hypertension Diabetes Dyslipidemia | 28.7 ± 5.1 71 (45%) 54 (34%) 99 (63%) |
| Liver disease etiology | |
| • NAFLD | 116 (74%) |
| • AIH | 32 (21%) |
| • Viral | 8 (5%) |
| Platelet count (×109/L) | 231 ± 76 |
| AST (U/L) | 39 (29–59) |
| ALT (U/L) GGT (U/L) | 49 (36–75) 81 (50–174) |
| Total bilirubin (mg/dL) Albumin (g/dL) INR | 0.6 (0.4–0.8) 4.4 (4.2–4.6) 1.0 (1.0–1.1) |
| Triglycerides (mg/dL) | 116 (80–166) |
| Total cholesterol (mg/dL) | 189 ± 41 |
| Histology | NAFLD (n = 116) | CH (n = 40) | |
|---|---|---|---|
| Feature | Grade/Stage | NASH-CRN Score | METAVIR Score |
| Inflammation | Grade 0 | 26 (22%) | 14 (34%) |
| Grade 1 | 58 (50%) | 8 (20%) | |
| Grade 2 | 28 (24%) | 9 (23%) | |
| Grade 3 | 4 (3%) | 9 (23%) | |
| Fibrosis | Stage 0 | 26 (22%) | 16 (40%) |
| Stage 1 | 28 (24%) | 9 (23%) | |
| Stage 2 | 26 (22%) | 8 (20%) | |
| Stage 3 | 18 (16%) | 5 (12%) | |
| Stage 4 | 18 (16%) | 2 (5%) | |
| Etiology | Histology Grade | I-Score (%) | p-Value | C-Score (foci/mm2) | p-Value |
|---|---|---|---|---|---|
| NAFLD | NASH-CRN 0 METAVIR 0 | 0.6 ± 0.5 | 0.23 | 2.7 ± 1.8 | 0.38 |
| CH | 0.4 ± 0.3 | 2.2 ± 1.6 | |||
| NAFLD | NASH-CRN 1 METAVIR 1 | 1.5 ± 1.2 | 0.58 | 6.8 ± 3.8 | 0.56 |
| CH | 1.8 ± 2.1 | 5.9 ± 3.0 | |||
| NAFLD | NASH-CRN 2 METAVIR 2 | 1.9 ± 1.4 | 0.005 | 8.5 ± 4.8 | 0.015 |
| CH | 5.4 ± 2.8 | 20.3 ± 11.5 | |||
| NAFLD | NASH-CRN 3 METAVIR 3 | 3.4 ± 2.1 | 0.04 | 12.5 ± 11.1 | 0.06 |
| CH | 6.6 ± 2.4 | 29.8 ± 18.6 |
| Etiology | Grades | I-Score Cut-Off (%) | I-Score AUC (95%CI) | C-Score Cut-Off (foci/mm2) | C-Score AUC (95%CI) | DeLong p-Value |
|---|---|---|---|---|---|---|
| NAFLD | NASH-CRN ≥ 1 METAVIR ≥ 1 | >0.6 | 0.83 (0.74–0.92) | >4.5 | 0.87 (0.80–0.94) | 0.20 |
| CH | >0.9 | 0.96 (0.92–1.00) | >4.4 | 0.96 (0.92–1.00) | 0.99 | |
| NAFLD | NASH-CRN ≥ 2 METAVIR ≥ 2 | >0.9 | 0.72 (0.62–0.82) | >6.6 | 0.72 (0.61–0.82) | 0.94 |
| CH | >2.0 | 0.97 (0.92–1.00) | >9.6 | 0.99 (0.97–1.00) | 0.22 | |
| NAFLD | NASH-CRN ≥ 3 METAVIR ≥ 3 | >2.2 | 0.83 (0.64–0.99) | >7.4 | 0.75 (0.56–0.93) | 0.16 |
| CH | >4.4 | 0.86 (0.74–0.97) | >12.3 | 0.91 (0.82–0.99) | 0.27 |
| Digital Pathology | NAFLD (n = 116) | Chronic Hepatitis (n = 40) | ||||
|---|---|---|---|---|---|---|
| Measurements | Simple Steatosis | NASH | p-Value | None/Low | Moderate/ Severe | p-Value |
| I-score (%) | 1.2 ± 1.3 | 1.6 ± 1.3 | 0.08 | 0.9 ± 1.4 | 6.0 ± 2.6 | <0.001 |
| C-score (foci/mm2) | 4.6 ± 3.9 | 7.9 ± 4.8 | <0.001 | 3.6 ± 2.8 | 25.1 ± 15.8 | <0.001 |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Parameter | OR (95%CI) | p-Value | OR (95%CI) | p-Value |
| AST 1 | 1.02 (1.0–1.04) | 0.025 | 1.01 (0.99–1.03) | 0.28 |
| ALT | 1.01 (0.99–1.02) | 0.37 | ||
| GGT | 1.0 (0.99–1.0) | 0.26 | ||
| Total bilirubin | 0.48 (0.17–1.39) | 0.17 | ||
| Albumin | 0.86 (0.33–2.23) | 0.74 | ||
| INR | 1.01 (0.97–1.04) | 0.29 | ||
| Platelets | 1.0 (0.99–1.01) | 0.75 | ||
| I-score 2 | 1.32 (0.97–1.80) | 0.08 | 1.71 (1.04–2.81) | 0.03 |
| C-score 2 | 1.24 (1.09–1.39) | <0.001 | 1.41 (1.16–1.70) | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marti-Aguado, D.; Fernández-Patón, M.; Alfaro-Cervello, C.; Mestre-Alagarda, C.; Bauza, M.; Gallen-Peris, A.; Merino, V.; Benlloch, S.; Pérez-Rojas, J.; Ferrández, A.; et al. Digital Pathology Enables Automated and Quantitative Assessment of Inflammatory Activity in Patients with Chronic Liver Disease. Biomolecules 2021, 11, 1808. https://doi.org/10.3390/biom11121808
Marti-Aguado D, Fernández-Patón M, Alfaro-Cervello C, Mestre-Alagarda C, Bauza M, Gallen-Peris A, Merino V, Benlloch S, Pérez-Rojas J, Ferrández A, et al. Digital Pathology Enables Automated and Quantitative Assessment of Inflammatory Activity in Patients with Chronic Liver Disease. Biomolecules. 2021; 11(12):1808. https://doi.org/10.3390/biom11121808
Chicago/Turabian StyleMarti-Aguado, David, Matías Fernández-Patón, Clara Alfaro-Cervello, Claudia Mestre-Alagarda, Mónica Bauza, Ana Gallen-Peris, Víctor Merino, Salvador Benlloch, Judith Pérez-Rojas, Antonio Ferrández, and et al. 2021. "Digital Pathology Enables Automated and Quantitative Assessment of Inflammatory Activity in Patients with Chronic Liver Disease" Biomolecules 11, no. 12: 1808. https://doi.org/10.3390/biom11121808