Regulation of Poly(ADP-Ribose) Polymerase 1 Activity by Y-Box-Binding Protein 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Proteins and Reagents
2.2. Preparation of DNA Duplexes and Plasmid DNA
2.3. Preparation of Mononucleosome Substrates
2.4. A Radioactive Assay of Protein PARylation by PARP1
2.5. AFM Experiments and Image Analysis
2.6. Fluorescence Anisotropy Measurements
2.7. Statistical Analysis
3. Results
3.1. Stimulation of PARP1 Activity by YB-1 is Affected by the Type of DNA Damage
3.2. YB-1 Modulates PARP1 Activation in the Presence of a Mononucleosome
3.3. Effects of Mg2+, Spermine, Histones, and YB-1 on PARP1 Activity In Vitro
3.4. YB-1 Decreases the Size of the PAR Polymer Produced by PARP1
3.5. YB-1 Modulates PAR Synthesis Activity in the HeLa Cell Extract
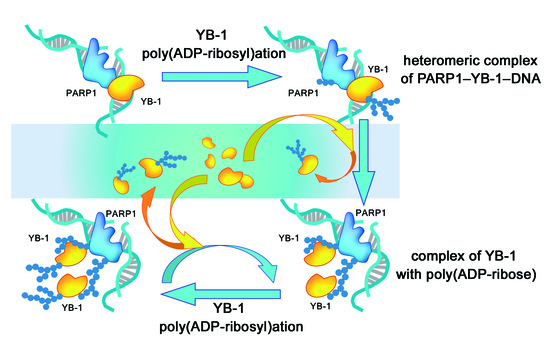
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef] [Green Version]
- Amé, J.C.; Spenlehauer, C.; De Murcia, G. The PARP superfamily. BioEssays 2004, 26, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Hottiger, M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015, 84, 227–263. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.S.; Poirier, G.G.; Lindahl, T. Dual Function for Poly(ADP-ribose) Synthesis in Response to DNA Strand Breakage. Biochemistry 1994, 33, 7099–7106. [Google Scholar] [CrossRef]
- Lindahl, T.; Satoh, M.S.; Poirier, G.G.; Klungland, A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995, 20, 405–411. [Google Scholar] [CrossRef]
- Amé, J.-C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Höger, T.; Murcia, J.M.D.; De Murcia, G. PARP-2, A Novel Mammalian DNA Damage-dependent Poly(ADP-ribose) Polymerase. J. Boil. Chem. 1999, 274, 17860–17868. [Google Scholar] [CrossRef] [Green Version]
- Leppard, J.B.; Dong, Z.; Mackey, Z.B.; Tomkinson, A.E. Physical and Functional Interaction between DNA Ligase IIIα and Poly(ADP-Ribose) Polymerase 1 in DNA Single-Strand Break Repair. Mol. Cell. Boil. 2003, 23, 5919–5927. [Google Scholar] [CrossRef] [Green Version]
- Tallis, M.; Morra, R.; Barkauskaite, E.; Ahel, I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma 2013, 123, 79–90. [Google Scholar] [CrossRef]
- Hanzlikova, H.; Gittens, W.; Krejcikova, K.; Zeng, Z.; Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017, 45, 2546–2557. [Google Scholar] [CrossRef]
- Ogata, N.; Ueda, K.; Kawaichi, M.; Hayaishi, O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Boil. Chem. 1981, 256, 4135–4137. [Google Scholar]
- Shieh, W.M.; Amé, J.C.; Wilson, M.V.; Wang, Z.Q.; Koh, D.W.; Jacobson, M.K.; Jacobson, E.L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Boil. Chem. 1998, 273, 30069–30072. [Google Scholar] [CrossRef] [Green Version]
- Vivelo, C.A.; Wat, R.; Agrawal, C.; Tee, H.Y.; Leung, A.K. ADPriboDB: The database of ADP-ribosylated proteins. Nucleic Acids Res. 2017, 45, 6254. [Google Scholar] [CrossRef] [Green Version]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) Binds to Specific Domains in DNA Damage Checkpoint Proteins. J. Boil. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef] [Green Version]
- Gagné, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jungmichel, S.; Rosenthal, F.; Altmeyer, M.; Lukas, J.; Hottiger, M.O.; Nielsen, M.L. Proteome-wide Identification of Poly(ADP-Ribosyl)ation Targets in Different Genotoxic Stress Responses. Mol. Cell 2013, 52, 272–285. [Google Scholar] [CrossRef] [Green Version]
- Gagné, J.P.; Pic, É.; Isabelle, M.; Krietsch, J.; Éthier, C.; Paquet, É.; Kelly, I.; Boutin, M.; Moon, K.M.; Foster, L.J.; et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012, 40, 7788–7805. [Google Scholar] [CrossRef] [Green Version]
- Hiroshi, S.; Toshio, M.; Fumio, I.; Kunio, Y.; Shunsuke, I. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene 1988, 73, 499–507. [Google Scholar] [CrossRef]
- Didier, D.K.; Schiffenbauer, J.; Woulfe, S.L.; Zacheis, M.; Schwartz, B.D. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA 1988, 85, 7322–7326. [Google Scholar] [CrossRef] [Green Version]
- Alemasova, E.E.; Moor, N.A.; Naumenko, K.N.; Kutuzov, M.M.; Sukhanova, M.V.; Pestryakov, P.E.; Lavrik, O.I. Y-box-binding protein 1 as a non-canonical factor of base excision repair. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2016, 1864, 1631–1640. [Google Scholar] [CrossRef]
- Das, S.; Chattopadhyay, R.; Bhakat, K.K.; Boldogh, I.; Kohno, K.; Prasad, R.; Wilson, S.H.; Hazra, T.K. Stimulation of NEIL2-mediated Oxidized Base Excision Repair via YB-1 Interaction during Oxidative Stress. J. Boil. Chem. 2007, 282, 28474–28484. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, R.; Das, S.; Maiti, A.K.; Boldogh, I.; Xie, J.; Hazra, T.K.; Kohno, K.; Mitra, S.; Bhakat, K.K. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol. Cell. Biol. 2008, 28, 7066–7080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guay, D.; Garand, C.; Reddy, S.; Schmutte, C.; Lebel, M. The human endonuclease III enzyme is a relevant target to potentiate cisplatin cytotoxicity in Y-box-binding protein-1 overexpressing tumor cells. Cancer Sci. 2008, 99, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Mordovkina, D.A.; Lyabin, D.N.; Smolin, E.A.; Sogorina, E.M.; Ovchinnikov, L.P.; Eliseeva, I.A. Y-Box Binding Proteins in mRNP Assembly, Translation, and Stability Control. Biomolecules 2020, 10, 591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koike, K.; Uchiumi, T.; Ohga, T.; Toh, S.; Wada, M.; Kohno, K.; Kuwano, M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997, 417, 390–394. [Google Scholar] [CrossRef]
- Ohga, T.; Uchiumi, T.; Makino, Y.; Koike, K.; Wada, M.; Kuwano, M.; Kohno, K. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Boil. Chem. 1998, 273, 5997–6000. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.B.; Ma, W.; Valova, V.A.; Algie, M.; Harfoot, R.; Woolley, A.G.; Robinson, P.J.; Braithwaite, A.W. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene 2009, 29, 403–410. [Google Scholar] [CrossRef] [Green Version]
- Kosnopfel, C.; Sinnberg, T.; Schittek, B. Y-box binding protein 1 – A prognostic marker and target in tumour therapy. Eur. J. Cell Boil. 2014, 93, 61–70. [Google Scholar] [CrossRef]
- Ise, T.; Nagatani, G.; Imamura, T.; Kato, K.; Takano, H.; Nomoto, M.; Izumi, H.; Ohmori, H.; Okamoto, T.; Ohga, T.; et al. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999, 59, 342–346. [Google Scholar]
- Marenstein, D.R.; Ocampo, M.T.A.; Chan, M.K.; Altamirano, A.; Basu, A.K.; Boorstein, R.J.; Cunningham, R.P.; Teebor, G.W. Stimulation of Human Endonuclease III by Y Box-binding Protein 1 (DNA-binding Protein B): INTERACTION BETWEEN A BASE EXCISION REPAIR ENZYME AND A TRANSCRIPTION FACTOR. J. Boil. Chem. 2001, 276, 21242–21249. [Google Scholar] [CrossRef] [Green Version]
- Gaudreault, I.; Guay, D.; Lebel, M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004, 32, 316–327. [Google Scholar] [CrossRef] [Green Version]
- Alemasova, E.E.; Pestryakov, P.E.; Sukhanova, M.V.; Kretov, D.A.; Moor, N.A.; Curmi, P.A.; Ovchinnikov, L.P.; Lavrik, O.I. Poly(ADP-ribosyl)ation as a new posttranslational modification of YB-1. Biochimie 2015, 119, 36–44. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Naumenko, K.N.; Kurgina, T.A.; Anarbaev, R.O.; Lavrik, O.I. The multifunctional protein YB-1 potentiates PARP1 activity and decreases the efficiency of PARP1 inhibitors. Oncotarget 2018, 9, 23349–23365. [Google Scholar] [CrossRef] [Green Version]
- Palazzo, L.; Ahel, I. PARPs in genome stability and signal transduction: Implications for cancer therapy. Biochem. Soc. Trans. 2018, 46, 1681–1695. [Google Scholar] [CrossRef] [Green Version]
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Fu, K.; Hodgson, A.; Wier, E.M.; Wen, M.G.; Kamenyeva, O.; Xia, X.; Koo, L.Y.; Wan, F. Sam68 Is Required for DNA Damage Responses via Regulating Poly(ADP-ribosyl)ation. PLoS Boil. 2016, 14, e1002543. [Google Scholar] [CrossRef] [Green Version]
- Kai, M. Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 2016, 17, 310. [Google Scholar] [CrossRef] [Green Version]
- Dolfini, D.; Mantovani, R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013, 20, 676–685. [Google Scholar] [CrossRef] [Green Version]
- Curtin, N.J. The Development of Rucaparib/Rubraca®: A Story of the Synergy Between Science and Serendipity. Cancers 2020, 12, 564. [Google Scholar] [CrossRef] [Green Version]
- Kutuzov, M.M.; Kurgina, T.A.; Belousova, E.A.; Khodyreva, S.N.; Lavrik, O.I. Optimization of nucleosome assembly from histones and model DNAs and estimation of the reconstitution efficiency. Biopolym. Cell 2019, 35, 91–98. [Google Scholar] [CrossRef]
- Nilsen, T.W. Preparation of Nuclear Extracts from HeLa Cells. Cold Spring Harb. Protoc. 2013, 2013, 579–583. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Naumenko, K.N.; Pestryakov, P.E.; Lavrik, O.I. Production, purification of the recombinant analog of Y-box-binding protein and its interaction with poly (ADP-ribose), RNA, single-and double-stranded DNAs. Biopolym. Cell 2017, 33, 214–220. [Google Scholar] [CrossRef] [Green Version]
- Sukhanova, M.V.; Khodyreva, S.N.; Lavrik, O.I. Poly(ADP-ribose) Polymerase-1 Inhibits Strand-Displacement Synthesis of DNA Catalyzed by DNA Polymerase β. Biochemistry 2004, 69, 558–568. [Google Scholar] [CrossRef]
- Lindahl, T.; Andersson, A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 1972, 11, 3618–3623. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Sukhanova, M.V.; Abrakhi, S.; Joshi, V.; Pastré, D.; Kutuzov, M.M.; Anarbaev, R.O.; Curmi, P.A.; Hamon, L.; Lavrik, O.I. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2015, 44, e60. [Google Scholar] [CrossRef] [Green Version]
- Révet, B. Short unligated sticky ends enable the observation of circularised DNA by atomic force and electron microscopies. Nucleic Acids Res. 1998, 26, 2092–2097. [Google Scholar] [CrossRef]
- Eliseeva, I.A.; Kim, E.R.; Guryanov, S.G.; Ovchinnikov, L.P.; Lyabin, D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef]
- Marsischky, G.T.; Wilson, B.A.; Collier, R.J. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Boil. Chem. 1995, 270, 3247–3254. [Google Scholar] [CrossRef] [Green Version]
- Eustermann, S.; Wu, W.-F.; Langelier, M.-F.; Yang, J.-C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754. [Google Scholar] [CrossRef] [Green Version]
- Wolffe, A.P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. BioEssays 1994, 16, 245–251. [Google Scholar] [CrossRef]
- Tanabe, Y.; Nagatoishi, S.; Tsumoto, K. Thermodynamic characterization of the interaction between the human Y-box binding protein YB-1 and nucleic acids. Mol. BioSyst. 2015, 11, 2441–2448. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Thonmas, J.O. Chromatin Structure: Oligomers of the Histones. Science 1974, 184, 865–868. [Google Scholar] [CrossRef]
- Arents, G.; Burlingame, R.W.; Wang, B.C.; Love, W.E.; Moudrianakis, E.N. The nucleosomal core histone octamer at 3.1 A resolution: A tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 1991, 88, 10148–10152. [Google Scholar] [CrossRef] [Green Version]
- Pinnola, A.; Naumova, N.; Shah, M.; Tulin, A.V. Nucleosomal Core Histones Mediate Dynamic Regulation of Poly(ADP-ribose) Polymerase 1 Protein Binding to Chromatin and Induction of Its Enzymatic Activity. J. Boil. Chem. 2007, 282, 32511–32519. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef] [Green Version]
- Clark, N.J.; Kramer, M.; Muthurajan, U.M.; Luger, K. Alternative Modes of Binding of Poly(ADP-ribose) Polymerase 1 to Free DNA and Nucleosomes. J. Boil. Chem. 2012, 287, 32430–32439. [Google Scholar] [CrossRef] [Green Version]
- Kurgina, T.; Anarbaev, R.; Sukhanova, M.; Lavrik, O.I. A rapid fluorescent method for the real-time measurement of poly(ADP-ribose) polymerase 1 activity. Anal. Biochem. 2018, 545, 91–97. [Google Scholar] [CrossRef]
- Krüger, A.; Bürkle, A.; Hauser, K.; Mangerich, A. Real-time monitoring of PARP1-dependent PARylation by ATR-FTIR spectroscopy. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Kreimeyer, A.; Wielckens, K.; Adamietz, P.; Hilz, H. DNA repair-associated ADP-ribosylation in vivo. Modification of histone H1 differs from that of the principal acceptor proteins. J. Boil. Chem. 1984, 259, 890–896. [Google Scholar]
- Kun, E.; Kirsten, E.; Mendeleyev, J.; Ordahl, C.P. Regulation of the Enzymatic Catalysis of Poly(ADP-ribose) Polymerase by dsDNA, Polyamines, Mg2+, Ca2+, Histones H1and H3, and ATP†. Biochemistry 2004, 43, 210–216. [Google Scholar] [CrossRef]
- Masaoka, A.; Gassman, N.R.; Kedar, P.S.; Prasad, R.; Hou, E.W.; Horton, J.K.; Bustin, M.; Wilson, S.H. HMGN1 Protein Regulates Poly(ADP-ribose) Polymerase-1 (PARP-1) Self-PARylation in Mouse Fibroblasts*. J. Boil. Chem. 2012, 287, 27648–27658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshihara, K.; Hashida, T.; Tanaka, Y.; Ohgushi, H.; Yoshihara, H.; Kamiya, T. Bovine thymus poly(adenosine diphosphate ribose) polymerase. J. Boil. Chem. 1978, 253, 6459–6466. [Google Scholar]
- Tanaka, Y.; Hashida, T.; Yoshihara, H.; Yoshihara, K. Bovine thymus poly(ADP-ribose) polymerase histone-dependent and Mg2+-dependent reaction. J. Boil. Chem. 1979, 254, 12433–12438. [Google Scholar]
- Alvarez-Gonzalez, R. 3′-Deoxy-NAD+ as a substrate for poly(ADP-ribose)polymerase and the reaction mechanism of poly(ADP-ribose) elongation. J. Boil. Chem. 1988, 263, 17690–17696. [Google Scholar]
- Brochu, G.; Duchaine, C.; Thibeault, L.; Lagueux, J.; Shah, G.M.; Poirier, G.G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1994, 1219, 342–350. [Google Scholar] [CrossRef]
- Bonicalzi, M.E.; Haince, J.F.; Droit, A.; Poirier, G.G. Regulation of poly (ADP-ribose) metabolism by poly (ADP-ribose) glycohydrolase: Where and when? Cell. Mol. Life Sci. 2005, 62, 739–750. [Google Scholar] [CrossRef]
- Lonskaya, I.; Potaman, V.N.; Shlyakhtenko, L.S.; Oussatcheva, E.A.; Lyubchenko, Y.L.; Soldatenkov, V.A. Regulation of Poly(ADP-ribose) Polymerase-1 by DNA Structure-specific Binding. J. Boil. Chem. 2005, 280, 17076–17083. [Google Scholar] [CrossRef] [Green Version]
- Langelier, M.F.; Riccio, A.A.; Pascal, J.M. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014, 42, 7762–7775. [Google Scholar] [CrossRef] [Green Version]
- Teloni, F.; Altmeyer, M. Readers of poly(ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 2015, 44, 993–1006. [Google Scholar] [CrossRef] [Green Version]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Boil. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Masson, M.; Niedergang, C.; Schreiber, V.; Muller, S.; Murcia, J.M.-D.; De Murcia, G. XRCC1 Is Specifically Associated with Poly(ADP-Ribose) Polymerase and Negatively Regulates Its Activity following DNA Damage. Mol. Cell. Boil. 1998, 18, 3563–3571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs-Seymour, I.; Fontana, P.; Rack, J.G.M.; Ahel, I. HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell 2016, 62, 432–442. [Google Scholar] [CrossRef]
- Maurya, P.K.; Mishra, A.; Yadav, B.S.; Singh, S.; Kumar, P.; Chaudhary, A.; Srivastava, S.; Murugesan, S.N.; Mani, A. Role of Y Box Protein-1 in cancer: As potential biomarker and novel therapeutic target. J. Cancer 2017, 8, 1900–1907. [Google Scholar] [CrossRef] [Green Version]
- Bargou, R.C.; Jürchott, K.; Wagener, C.; Bergmann, S.; Metzner, S.; Bommert, K.; Mapara, M.Y.; Winzer, K.J.; Dietel, M.; Dörken, B.; et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat. Med. 1997, 3, 447–450. [Google Scholar] [CrossRef]
- Fischbach, A.; Krüger, A.; Hampp, S.; Assmann, G.; Rank, L.; Hufnagel, M.; Stöckl, M.T.; Fischer, J.M.; Veith, S.; Rossatti, P.; et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2017, 46, 804–822. [Google Scholar] [CrossRef] [Green Version]












| Name | Oligodeoxynucleotide Sequences (5′→3′) |
|---|---|
| ss32 | 5′-GGCGATTAAGTTGGGAAACGTCAGGGTCTTCC-3′ |
| ds55 | 5′-CGGTATCCACCAGGTCTGAGACAACGATGAAGCCCAAGCCAGATGAAATGTAGTC-3′ 3′-GCCATAGGTGGTCCTGACTCTGTTGCTACTTCGGGTTCGGTCTACTTTACATCAG-5′ |
| mm55 | 5′-CGGTATCCACCAGGTCTGAGACAACGATGAAGCCCAAGCCAGATGAAATGTAGTC-3′ 3′-GCCATAGGTGGTCCTGACGCTGTTGCTACTTCGGGTTCGGTCTACTTTACATCAG-5′ |
| boldfaced nucleotides indicate the position of the A–G mismatch | |
| Bubble(5) | 5′-CGGTATCCACCAGGTCACUCTCAACGATGAAGCCCAAGCCAGATGAAATGTAGTC-3′ 3′-GCCATAGGTGGTCCTGACGCTGTTGCTACTTCGGGTTCGGTCTACTTTACATCAG-5′ |
| boldfaced nucleotides indicate the position of the 5 nt bubble | |
| Bubble(17) | 5′-CGGTATCCACGTCCATACUCTGTGTTGTGAAGCCCAAGCCAGATGAAATGTAGTC-3′ 3′-GCCATAGGTGGTCCAGACGCTGTTGCTACTTCGGGTTCGGTCTACTTTACATCAG-5′ |
| boldfaced nucleotides indicate the position of the 17 nt bubble | |
| Nick | 3′-OH\ /5′-Phosphate 5′-GGCGATAAAGTTGGGAAACGTCAGGGTCTTCC-3′ 3′-CCGCTATTTCAACCCTTTGCAGTCCCAGAAGG-5′ |
| Gap (one-nucleotide) | 3′-OH\ /5′-Phosphate 5′-GGCGATAAAGTTGGG AACGTCAGGGTCTTCC-3′ 3′-CCGCTATTTCAACCCTTTGCAGTCCCAGAAGG-5′ |
| 3′-Recessed | 5′-GGCGATAAAGTTGGG-3′ 3′-CCGCTATTTCAACCCTTTGCAGTCCCAGAAGG-5′ |
| Dumbbell | T-T-GCTTGAAGGCGCTTCGAAGACGG-T-T | | T-T-CGAACTTCCGCGAAGCTTCTGCC-T-T |
| Gap dumbbell (one-nucleotide) | 3′-OH\ /5′-Phosphate T-T-GCTTGAAGGCG TTCGAAGACGG-T-T | | T-T-CGAACTTCCGCGAAGCTTCTGCC-T-T |
| Nick dumbbell | 3′-OH\ /5′-Phosphate T-T-GCTTGAAGGCGCTTCGAAGACGG-T-T | | T-T-CGAACTTCCGCGAAGCTTCTGCC-T-T |
| Name; Sequence in the Circle | Scheme |
|---|---|
| 147 bp DNA 5′-ACCCCAGGGACUTGAAGTAATAA-3′ 3′-TGGGGTCCCTGAACTTCATTATT-5′ |  |
| 147 bp DNA (gap) OH\ /Phosphate 5′-ACCCCAGGGAC TGAAGTAATAA-3′ 3′-TGGGGTCCCTGAACTTCATTATT-5′ |  |
| Mononucleosome 5′-ACCCCAGGGACUTGAAGTAATAA-3′ 3′-TGGGGTCCCTGAACTTCATTATT-5′ |  |
| Mononucleosome (gap) OH\ /Phosphate 5′-ACCCCAGGGAC TGAAGTAATAA-3′ 3′-TGGGGTCCCTGAACTTCATTATT-5′ |  |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumenko, K.N.; Sukhanova, M.V.; Hamon, L.; Kurgina, T.A.; Alemasova, E.E.; Kutuzov, M.M.; Pastré, D.; Lavrik, O.I. Regulation of Poly(ADP-Ribose) Polymerase 1 Activity by Y-Box-Binding Protein 1. Biomolecules 2020, 10, 1325. https://doi.org/10.3390/biom10091325
Naumenko KN, Sukhanova MV, Hamon L, Kurgina TA, Alemasova EE, Kutuzov MM, Pastré D, Lavrik OI. Regulation of Poly(ADP-Ribose) Polymerase 1 Activity by Y-Box-Binding Protein 1. Biomolecules. 2020; 10(9):1325. https://doi.org/10.3390/biom10091325
Chicago/Turabian StyleNaumenko, Konstantin N., Mariya V. Sukhanova, Loic Hamon, Tatyana A. Kurgina, Elizaveta E. Alemasova, Mikhail M. Kutuzov, David Pastré, and Olga I. Lavrik. 2020. "Regulation of Poly(ADP-Ribose) Polymerase 1 Activity by Y-Box-Binding Protein 1" Biomolecules 10, no. 9: 1325. https://doi.org/10.3390/biom10091325