Defining the Protein Seeds of Neurodegeneration using Real-Time Quaking-Induced Conversion Assays
Abstract
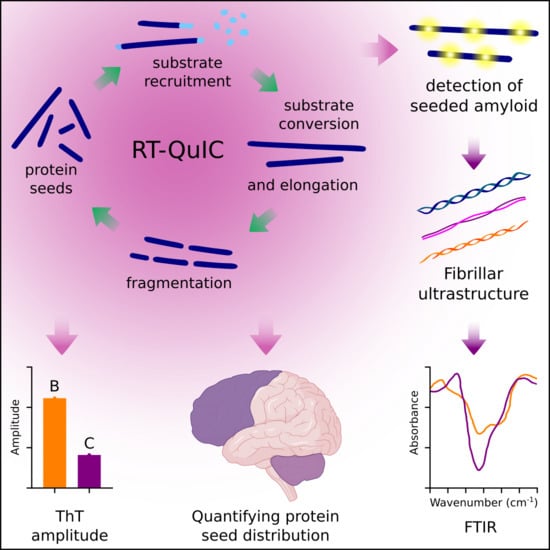
:1. Introduction
2. RT-QuIC Assays as Diagnostic Tools
2.1. Tau
2.2. αSynuclein
3. Application of RT-QuIC Assays to Understand Characteristics of Protein Seeds
Exploiting Strains for Diagnosis and Dissecting Disease-Related Mechanisms
4. Defining Protein Seed Distributions and Networks
4.1. Seeding Activity Assessments of Prion Propagation and Transmission
4.2. The Networks of Tau and αSyn Seeds
5. RT-QuIC Assays to Determine the Structural Modalities of Seeds and to Test Anti-amyloid Drugs and Disinfectants
Advantages, Limitations, and Further Development of RT-QuIC Assays
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moussaud, S.; Jones, D.R.; Moussaud-Lamodiere, E.L.; Delenclos, M.; Ross, O.A.; McLean, P.J. Alpha-synuclein and tau: Teammates in neurodegeneration? Mol. Neurodegener. 2014, 9, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visanji, N.P.; Lang, A.E.; Kovacs, G.G. Beyond the synucleinopathies: Alpha synuclein as a driving force in neurodegenerative comorbidities. Transl. Neurodegener. 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caughey, B.; Kraus, A. Transmissibility versus Pathogenicity of Self-Propagating Protein Aggregates. Viruses 2019, 11, 1044. [Google Scholar] [CrossRef] [Green Version]
- Silveira, J.R.; Raymond, G.J.; Hughson, A.G.; Race, R.E.; Sim, V.L.; Hayes, S.F.; Caughey, B. The most infectious prion protein particles. Nature 2005, 437, 257–261. [Google Scholar] [CrossRef] [Green Version]
- Caughey, B.; Kocisko, D.A.; Raymond, G.J.; Lansbury, P.T., Jr. Aggregates of scrapie-associated prion protein induce the cell-free conversion of protease-sensitive prion protein to the protease-resistant state. Chem. Biol. 1995, 2, 807–817. [Google Scholar] [CrossRef] [Green Version]
- Chesebro, B.; Trifilo, M.; Race, R.; Meade-White, K.; Teng, C.; LaCasse, R.; Raymond, L.; Favara, C.; Baron, G.; Priola, S.; et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 2005, 308, 1435–1439. [Google Scholar] [CrossRef] [Green Version]
- Terry, C.; Harniman, R.L.; Sells, J.; Wenborn, A.; Joiner, S.; Saibil, H.R.; Miles, M.J.; Collinge, J.; Wadsworth, J.D.F. Structural features distinguishing infectious ex vivo mammalian prions from non-infectious fibrillar assemblies generated in vitro. Sci. Rep. 2019, 9, 376. [Google Scholar] [CrossRef] [Green Version]
- Vascellari, S.; Orru, C.D.; Hughson, A.G.; King, D.; Barron, R.; Wilham, J.M.; Baron, G.S.; Race, B.; Pani, A.; Caughey, B. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS ONE 2012, 7, e48969. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Haldiman, T.; Surewicz, K.; Cohen, Y.; Chen, W.; Blevins, J.; Sy, M.S.; Cohen, M.; Kong, Q.; Telling, G.C.; et al. Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrP(C). PLoS Pathog. 2012, 8, e1002835. [Google Scholar] [CrossRef]
- Kraus, A.; Raymond, G.J.; Race, B.; Campbell, K.J.; Hughson, A.G.; Anson, K.J.; Raymond, L.D.; Caughey, B. PrP P102L and Nearby Lysine Mutations Promote Spontaneous In Vitro Formation of Transmissible Prions. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Groveman, B.R.; Raymond, G.J.; Campbell, K.J.; Race, B.; Raymond, L.D.; Hughson, A.G.; Orru, C.D.; Kraus, A.; Phillips, K.; Caughey, B. Role of the central lysine cluster and scrapie templating in the transmissibility of synthetic prion protein aggregates. PLoS Pathog. 2017, 13, e1006623. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, X.; Orru, C.D.; Groveman, B.R.; Surewicz, K.; Abskharon, R.; Imamura, M.; Yokoyama, T.; Kim, Y.S.; Vander Stel, K.J.; et al. Self-propagating, protease-resistant, recombinant prion protein conformers with or without in vivo pathogenicity. PLoS Pathog. 2017, 13, e1006491. [Google Scholar] [CrossRef] [PubMed]
- Makarava, N.; Kovacs, G.G.; Savtchenko, R.; Alexeeva, I.; Budka, H.; Rohwer, R.G.; Baskakov, I.V. Genesis of mammalian prions: From non-infectious amyloid fibrils to a transmissible prion disease. PLoS Pathog. 2011, 7, e1002419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond, G.J.; Race, B.; Hollister, J.R.; Offerdahl, D.K.; Moore, R.A.; Kodali, R.; Raymond, L.D.; Hughson, A.G.; Rosenke, R.; Long, D.; et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J. Virol. 2012, 86, 11763–11778. [Google Scholar] [CrossRef] [Green Version]
- Legname, G.; Baskakov, I.V.; Nguyen, H.O.; Riesner, D.; Cohen, F.E.; DeArmond, S.J.; Prusiner, S.B. Synthetic mammalian prions. Science 2004, 305, 673–676. [Google Scholar] [CrossRef]
- Vazquez-Fernandez, E.; Vos, M.R.; Afanasyev, P.; Cebey, L.; Sevillano, A.M.; Vidal, E.; Rosa, I.; Renault, L.; Ramos, A.; Peters, P.J.; et al. The Structural Architecture of an Infectious Mammalian Prion Using Electron Cryomicroscopy. PLoS Pathog. 2016, 12, e1005835. [Google Scholar] [CrossRef]
- Groveman, B.R.; Dolan, M.A.; Taubner, L.M.; Kraus, A.; Wickner, R.B.; Caughey, B. Parallel in-register intermolecular beta-sheet architectures for prion-seeded prion protein (PrP) amyloids. J. Biol. Chem. 2014, 289, 24129–24142. [Google Scholar] [CrossRef] [Green Version]
- Spagnolli, G.; Rigoli, M.; Orioli, S.; Sevillano, A.M.; Faccioli, P.; Wille, H.; Biasini, E.; Requena, J.R. Full atomistic model of prion structure and conversion. PLoS Pathog. 2019, 15, e1007864. [Google Scholar] [CrossRef] [Green Version]
- Safar, J.; Wille, H.; Itri, V.; Groth, D.; Serban, H.; Torchia, M.; Cohen, F.E.; Prusiner, S.B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998, 4, 1157–1165. [Google Scholar] [CrossRef]
- Bessen, R.A.; Marsh, R.F. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 1992, 66, 2096–2101. [Google Scholar] [CrossRef] [Green Version]
- Telling, G.C.; Parchi, P.; DeArmond, S.J.; Cortelli, P.; Montagna, P.; Gabizon, R.; Mastrianni, J.; Lugaresi, E.; Gambetti, P.; Prusiner, S.B. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 1996, 274, 2079–2082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narasimhan, S.; Guo, J.L.; Changolkar, L.; Stieber, A.; McBride, J.D.; Silva, L.V.; He, Z.; Zhang, B.; Gathagan, R.J.; Trojanowski, J.Q.; et al. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. 2017, 37, 11406–11423. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.L.; Lee, V.M. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011, 286, 15317–15331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clavaguera, F.; Lavenir, I.; Falcon, B.; Frank, S.; Goedert, M.; Tolnay, M. “Prion-like” templated misfolding in tauopathies. Brain Pathol. 2013, 23, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Luk, K.C.; Kehm, V.; Carroll, J.; Zhang, B.; O’Brien, P.; Trojanowski, J.Q.; Lee, V.M. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 2012, 338, 949–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bousset, L.; Pieri, L.; Ruiz-Arlandis, G.; Gath, J.; Jensen, P.H.; Habenstein, B.; Madiona, K.; Olieric, V.; Bockmann, A.; Meier, B.H.; et al. Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 2013, 4, 2575. [Google Scholar] [CrossRef] [Green Version]
- Peelaerts, W.; Bousset, L.; Van der Perren, A.; Moskalyuk, A.; Pulizzi, R.; Giugliano, M.; Van den Haute, C.; Melki, R.; Baekelandt, V. alpha-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 2015, 522, 340–344. [Google Scholar] [CrossRef]
- Kaufman, S.K.; Sanders, D.W.; Thomas, T.L.; Ruchinskas, A.J.; Vaquer-Alicea, J.; Sharma, A.M.; Miller, T.M.; Diamond, M.I. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo. Neuron 2016, 92, 796–812. [Google Scholar] [CrossRef] [Green Version]
- Lavenir, I.; Passarella, D.; Masuda-Suzukake, M.; Curry, A.; Holton, J.L.; Ghetti, B.; Goedert, M. Silver staining (Campbell-Switzer) of neuronal alpha-synuclein assemblies induced by multiple system atrophy and Parkinson’s disease brain extracts in transgenic mice. Acta Neuropathol. Commun. 2019, 7, 148. [Google Scholar] [CrossRef]
- Holmes, B.B.; Furman, J.L.; Mahan, T.E.; Yamasaki, T.R.; Mirbaha, H.; Eades, W.C.; Belaygorod, L.; Cairns, N.J.; Holtzman, D.M.; Diamond, M.I. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, E4376–E4385. [Google Scholar] [CrossRef] [Green Version]
- Woerman, A.L.; Stohr, J.; Aoyagi, A.; Rampersaud, R.; Krejciova, Z.; Watts, J.C.; Ohyama, T.; Patel, S.; Widjaja, K.; Oehler, A.; et al. Propagation of prions causing synucleinopathies in cultured cells. Proc. Natl. Acad. Sci. USA 2015, 112, E4949–E4958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furman, J.L.; Vaquer-Alicea, J.; White, C.L., 3rd; Cairns, N.J.; Nelson, P.T.; Diamond, M.I. Widespread tau seeding activity at early Braak stages. Acta Neuropathol. 2017, 133, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.K.; Del Tredici, K.; Thomas, T.L.; Braak, H.; Diamond, M.I. Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer’s disease and PART. Acta Neuropathol. 2018, 136, 57–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saborio, G.P.; Permanne, B.; Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001, 411, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, R.; Moore, R.A.; Sim, V.L.; Hughson, A.G.; Dorward, D.W.; Onwubiko, H.A.; Priola, S.A.; Caughey, B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 2007, 4, 645–650. [Google Scholar] [CrossRef]
- Shahnawaz, M.; Mukherjee, A.; Pritzkow, S.; Mendez, N.; Rabadia, P.; Liu, X.; Hu, B.; Schmeichel, A.; Singer, W.; Wu, G.; et al. Discriminating alpha-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020, 578, 273–277. [Google Scholar] [CrossRef]
- Saa, P.; Castilla, J.; Soto, C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J. Biol. Chem. 2006, 281, 35245–35252. [Google Scholar] [CrossRef] [Green Version]
- Castilla, J.; Morales, R.; Saa, P.; Barria, M.; Gambetti, P.; Soto, C. Cell-free propagation of prion strains. EMBO J. 2008, 27, 2557–2566. [Google Scholar] [CrossRef] [Green Version]
- Strohaker, T.; Jung, B.C.; Liou, S.H.; Fernandez, C.O.; Riedel, D.; Becker, S.; Halliday, G.M.; Bennati, M.; Kim, W.S.; Lee, S.J.; et al. Structural heterogeneity of alpha-synuclein fibrils amplified from patient brain extracts. Nat. Commun. 2019, 10, 5535. [Google Scholar] [CrossRef] [Green Version]
- Shahnawaz, M.; Tokuda, T.; Waragai, M.; Mendez, N.; Ishii, R.; Trenkwalder, C.; Mollenhauer, B.; Soto, C. Development of a Biochemical Diagnosis of Parkinson Disease by Detection of alpha-Synuclein Misfolded Aggregates in Cerebrospinal Fluid. JAMA Neurol. 2017, 74, 163–172. [Google Scholar] [CrossRef]
- Salvadores, N.; Shahnawaz, M.; Scarpini, E.; Tagliavini, F.; Soto, C. Detection of misfolded Abeta oligomers for sensitive biochemical diagnosis of Alzheimer’s disease. Cell Rep. 2014, 7, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, R.M. Animal Virus Titration Techniques. In Techniques in Experimental Virology; Harris, R.J.C., Ed.; Academic Press Inc.: New York, NY, USA, 1964; pp. 183–186. [Google Scholar]
- Metrick, M.A., 2nd; do Carmo Ferreira, N.; Saijo, E.; Hughson, A.G.; Kraus, A.; Orru, C.; Miller, M.W.; Zanusso, G.; Ghetti, B.; Vendruscolo, M.; et al. Million-fold sensitivity enhancement in proteopathic seed amplification assays for biospecimens by Hofmeister ion comparisons. Proc. Natl. Acad. Sci. USA 2019, 116, 23029–23039. [Google Scholar] [CrossRef] [PubMed]
- Kraus, A.; Saijo, E.; Metrick, M.A., 2nd; Newell, K.; Sigurdson, C.J.; Zanusso, G.; Ghetti, B.; Caughey, B. Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 2019, 137, 585–598. [Google Scholar] [CrossRef] [Green Version]
- Saijo, E.; Ghetti, B.; Zanusso, G.; Oblak, A.; Furman, J.L.; Diamond, M.I.; Kraus, A.; Caughey, B. Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol. 2017, 133, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Saijo, E.; Metrick, M.A., 2nd; Koga, S.; Parchi, P.; Litvan, I.; Spina, S.; Boxer, A.; Rojas, J.C.; Galasko, D.; Kraus, A.; et al. 4-Repeat tau seeds and templating subtypes as brain and CSF biomarkers of frontotemporal lobar degeneration. Acta Neuropathol. 2019. [Google Scholar] [CrossRef]
- Orru, C.D.; Hughson, A.G.; Groveman, B.R.; Campbell, K.J.; Anson, K.J.; Manca, M.; Kraus, A.; Caughey, B. Factors That Improve RT-QuIC Detection of Prion Seeding Activity. Viruses 2016, 8, 140. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, R.; Satoh, K.; Sano, K.; Fuse, T.; Yamaguchi, N.; Ishibashi, D.; Matsubara, T.; Nakagaki, T.; Yamanaka, H.; Shirabe, S.; et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 2011, 17, 175–178. [Google Scholar] [CrossRef] [Green Version]
- Wilham, J.M.; Orru, C.D.; Bessen, R.A.; Atarashi, R.; Sano, K.; Race, B.; Meade-White, K.D.; Taubner, L.M.; Timmes, A.; Caughey, B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010, 6, e1001217. [Google Scholar] [CrossRef] [Green Version]
- Foutz, A.; Appleby, B.S.; Hamlin, C.; Liu, X.; Yang, S.; Cohen, Y.; Chen, W.; Blevins, J.; Fausett, C.; Wang, H.; et al. Diagnostic and prognostic value of human prion detection in cerebrospinal fluid. Ann. Neurol. 2017, 81, 79–92. [Google Scholar] [CrossRef]
- Green, A.J.E. RT-QuIC: A new test for sporadic CJD. Pract. Neurol. 2019, 19, 49–55. [Google Scholar] [CrossRef]
- Franceschini, A.; Baiardi, S.; Hughson, A.G.; McKenzie, N.; Moda, F.; Rossi, M.; Capellari, S.; Green, A.; Giaccone, G.; Caughey, B.; et al. High diagnostic value of second generation CSF RT-QuIC across the wide spectrum of CJD prions. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bongianni, M.; Orru, C.; Groveman, B.R.; Sacchetto, L.; Fiorini, M.; Tonoli, G.; Triva, G.; Capaldi, S.; Testi, S.; Ferrari, S.; et al. Diagnosis of Human Prion Disease Using Real-Time Quaking-Induced Conversion Testing of Olfactory Mucosa and Cerebrospinal Fluid Samples. JAMA Neurol. 2017, 74, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wrona, A.; Foutz, A.; Blevins, J.; Glisic, K.; Person, M.; Maddox, R.A.; Belay, E.D.; Schonberger, L.B.; Tatsuoka, C.; et al. Diagnosis of Prion Diseases by RT-QuIC Results in Improved Surveillance. Neurology 2020. [Google Scholar] [CrossRef] [PubMed]
- McGuire, L.I.; Peden, A.H.; Orru, C.D.; Wilham, J.M.; Appleford, N.E.; Mallinson, G.; Andrews, M.; Head, M.W.; Caughey, B.; Will, R.G.; et al. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 2012, 72, 278–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orru, C.D.; Groveman, B.R.; Hughson, A.G.; Zanusso, G.; Coulthart, M.B.; Caughey, B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. MBIO 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Orru, C.D.; Bongianni, M.; Tonoli, G.; Ferrari, S.; Hughson, A.G.; Groveman, B.R.; Fiorini, M.; Pocchiari, M.; Monaco, S.; Caughey, B.; et al. A test for Creutzfeldt-Jakob disease using nasal brushings. N. Engl. J. Med. 2014, 371, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Orru, C.D.; Soldau, K.; Cordano, C.; Llibre-Guerra, J.; Green, A.J.; Sanchez, H.; Groveman, B.R.; Edland, S.D.; Safar, J.G.; Lin, J.H.; et al. Prion Seeds Distribute throughout the Eyes of Sporadic Creutzfeldt-Jakob Disease Patients. MBIO 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Orru, C.D.; Yuan, J.; Appleby, B.S.; Li, B.; Li, Y.; Winner, D.; Wang, Z.; Zhan, Y.A.; Rodgers, M.; Rarick, J.; et al. Prion seeding activity and infectivity in skin samples from patients with sporadic Creutzfeldt-Jakob disease. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Mammana, A.; Baiardi, S.; Rossi, M.; Franceschini, A.; Donadio, V.; Capellari, S.; Caughey, B.; Parchi, P. Detection of prions in skin punch biopsies of Creutzfeldt-Jakob disease patients. Ann. Clin. Transl. Neurol. 2020, 7, 559–564. [Google Scholar] [CrossRef]
- Goedert, M.; Wischik, C.M.; Crowther, R.A.; Walker, J.E.; Klug, A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: Identification as the microtubule-associated protein tau. Proc. Natl. Acad. Sci. USA 1988, 85, 4051–4055. [Google Scholar] [CrossRef] [Green Version]
- Goedert, M.; Spillantini, M.G.; Potier, M.C.; Ulrich, J.; Crowther, R.A. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J. 1989, 8, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Falcon, B.; Zivanov, J.; Zhang, W.; Murzin, A.G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Newell, K.L.; Ghetti, B.; Goedert, M.; et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 2019, 568, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tarutani, A.; Newell, K.L.; Murzin, A.G.; Matsubara, T.; Falcon, B.; Vidal, R.; Garringer, H.J.; Shi, Y.; Ikeuchi, T.; et al. Novel tau filament fold in corticobasal degeneration. Nature 2020, 580, 283–287. [Google Scholar] [CrossRef]
- Arakhamia, T.; Lee, C.E.; Carlomagno, Y.; Duong, D.M.; Kundinger, S.R.; Wang, K.; Williams, D.; DeTure, M.; Dickson, D.W.; Cook, C.N.; et al. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 2020, 180, 633–644 e612. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. Ordered Assembly of Tau Protein and Neurodegeneration. Adv. Exp. Med. Biol. 2019, 1184, 3–21. [Google Scholar] [CrossRef]
- Crary, J.F.; Trojanowski, J.Q.; Schneider, J.A.; Abisambra, J.F.; Abner, E.L.; Alafuzoff, I.; Arnold, S.E.; Attems, J.; Beach, T.G.; Bigio, E.H.; et al. Primary age-related tauopathy (PART): A common pathology associated with human aging. Acta Neuropathol. 2014, 128, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Metrick, M.A., 2nd; Ferreira, N.D.C.; Saijo, E.; Kraus, A.; Newell, K.; Zanusso, G.; Vendruscolo, M.; Ghetti, B.; Caughey, B. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol. Commun. 2020, 8, 22. [Google Scholar] [CrossRef]
- Fairfoul, G.; McGuire, L.I.; Pal, S.; Ironside, J.W.; Neumann, J.; Christie, S.; Joachim, C.; Esiri, M.; Evetts, S.G.; Rolinski, M.; et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann. Clin. Transl. Neurol. 2016, 3, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Groveman, B.R.; Orru, C.D.; Hughson, A.G.; Raymond, L.D.; Zanusso, G.; Ghetti, B.; Campbell, K.J.; Safar, J.; Galasko, D.; Caughey, B. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol. Commun. 2018, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Kang, U.J.; Boehme, A.K.; Fairfoul, G.; Shahnawaz, M.; Ma, T.C.; Hutten, S.J.; Green, A.; Soto, C. Comparative study of cerebrospinal fluid alpha-synuclein seeding aggregation assays for diagnosis of Parkinson’s disease. Mov. Disord. 2019, 34, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Bongianni, M.; Ladogana, A.; Capaldi, S.; Klotz, S.; Baiardi, S.; Cagnin, A.; Perra, D.; Fiorini, M.; Poleggi, A.; Legname, G.; et al. alpha-Synuclein RT-QuIC assay in cerebrospinal fluid of patients with dementia with Lewy bodies. Ann. Clin. Transl. Neurol. 2019, 6, 2120–2126. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Candelise, N.; Baiardi, S.; Capellari, S.; Giannini, G.; Orru, C.D.; Antelmi, E.; Mammana, A.; Hughson, A.G.; Calandra-Buonaura, G.; et al. Ultrasensitive RT-QuIC assay with high sensitivity and specificity for Lewy body-associated synucleinopathies. Acta Neuropathol. 2020. [Google Scholar] [CrossRef]
- van Rumund, A.; Green, A.J.E.; Fairfoul, G.; Esselink, R.A.J.; Bloem, B.R.; Verbeek, M.M. alpha-Synuclein real-time quaking-induced conversion in the cerebrospinal fluid of uncertain cases of parkinsonism. Ann. Neurol. 2019, 85, 777–781. [Google Scholar] [CrossRef] [Green Version]
- Barone, D.A.; Henchcliffe, C. Rapid eye movement sleep behavior disorder and the link to alpha-synucleinopathies. Clin. Neurophysiol. 2018, 129, 1551–1564. [Google Scholar] [CrossRef]
- Coon, E.A.; Singer, W.; Low, P.A. Pure Autonomic Failure. Mayo Clin. Proc. 2019, 94, 2087–2098. [Google Scholar] [CrossRef] [Green Version]
- Garrido, A.; Fairfoul, G.; Tolosa, E.S.; Marti, M.J.; Green, A.; Barcelona, L.S.G. Alpha-synuclein RT-QuIC in cerebrospinal fluid of LRRK2-linked Parkinson’s disease. Ann. Clin. Transl. Neurol. 2019, 6, 1024–1032. [Google Scholar] [CrossRef] [Green Version]
- De Luca, C.M.G.; Elia, A.E.; Portaleone, S.M.; Cazzaniga, F.A.; Rossi, M.; Bistaffa, E.; De Cecco, E.; Narkiewicz, J.; Salzano, G.; Carletta, O.; et al. Efficient RT-QuIC seeding activity for alpha-synuclein in olfactory mucosa samples of patients with Parkinson’s disease and multiple system atrophy. Transl. Neurodegener. 2019, 8, 24. [Google Scholar] [CrossRef]
- Manne, S.; Kondru, N.; Jin, H.; Anantharam, V.; Huang, X.; Kanthasamy, A.; Kanthasamy, A.G. alpha-Synuclein real-time quaking-induced conversion in the submandibular glands of Parkinson’s disease patients. Mov. Disord. 2020, 35, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, T.R.; Holmes, B.B.; Furman, J.L.; Dhavale, D.D.; Su, B.W.; Song, E.S.; Cairns, N.J.; Kotzbauer, P.T.; Diamond, M.I. Parkinson’s disease and multiple system atrophy have distinct alpha-synuclein seed characteristics. J. Biol. Chem. 2019, 294, 1045–1058. [Google Scholar] [CrossRef] [Green Version]
- Schweighauser, M.; Shi, Y.; Tarutani, A.; Kametani, F.; Murzin, A.G.; Ghetti, B.; Matsubara, T.; Tomita, T.; Ando, T.; Hasegawa, K.; et al. Structures of alpha-synuclein filaments from multiple system atrophy. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Ubagai, K.; Fukuda, S.; Mori, T.; Takatsuki, H.; Taguchi, Y.; Kageyama, S.; Nishida, N.; Atarashi, R. Discrimination between L-type and C-type bovine spongiform encephalopathy by the strain-specific reactions of real-time quaking-induced conversion. Biochem. Biophys. Res. Commun. 2020, 526, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Orru, C.D.; Favole, A.; Corona, C.; Mazza, M.; Manca, M.; Groveman, B.R.; Hughson, A.G.; Acutis, P.L.; Caramelli, M.; Zanusso, G.; et al. Detection and discrimination of classical and atypical L-type bovine spongiform encephalopathy by real-time quaking-induced conversion. J. Clin. Microbiol. 2015, 53, 1115–1120. [Google Scholar] [CrossRef] [Green Version]
- Masujin, K.; Orru, C.D.; Miyazawa, K.; Groveman, B.R.; Raymond, L.D.; Hughson, A.G.; Caughey, B. Detection of Atypical H-Type Bovine Spongiform Encephalopathy and Discrimination of Bovine Prion Strains by Real-Time Quaking-Induced Conversion. J. Clin. Microbiol. 2016, 54, 676–686. [Google Scholar] [CrossRef] [Green Version]
- Orru, C.D.; Groveman, B.R.; Raymond, L.D.; Hughson, A.G.; Nonno, R.; Zou, W.; Ghetti, B.; Gambetti, P.; Caughey, B. Bank Vole Prion Protein as an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains. PLoS Pathog. 2015, 11, e1004983. [Google Scholar] [CrossRef]
- Bistaffa, E.; Vuong, T.T.; Cazzaniga, F.A.; Tran, L.; Salzano, G.; Legname, G.; Giaccone, G.; Benestad, S.L.; Moda, F. Use of different RT-QuIC substrates for detecting CWD prions in the brain of Norwegian cervids. Sci. Rep. 2019, 9, 18595. [Google Scholar] [CrossRef] [Green Version]
- Dickson, D.W.; Liu, W.; Hardy, J.; Farrer, M.; Mehta, N.; Uitti, R.; Mark, M.; Zimmerman, T.; Golbe, L.; Sage, J.; et al. Widespread alterations of alpha-synuclein in multiple system atrophy. Am. J. Pathol. 1999, 155, 1241–1251. [Google Scholar] [CrossRef]
- Candelise, N.; Schmitz, M.; Llorens, F.; Villar-Pique, A.; Cramm, M.; Thom, T.; da Silva Correia, S.M.; da Cunha, J.E.G.; Mobius, W.; Outeiro, T.F.; et al. Seeding variability of different alpha synuclein strains in synucleinopathies. Ann. Neurol. 2019, 85, 691–703. [Google Scholar] [CrossRef]
- Orru, C.D.; Hughson, A.G.; Race, B.; Raymond, G.J.; Caughey, B. Time course of prion seeding activity in cerebrospinal fluid of scrapie-infected hamsters after intratongue and intracerebral inoculations. J. Clin. Microbiol. 2012, 50, 1464–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chesebro, B.; Striebel, J.; Rangel, A.; Phillips, K.; Hughson, A.; Caughey, B.; Race, B. Early Generation of New PrPSc on Blood Vessels after Brain Microinjection of Scrapie in Mice. MBIO 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Manca, M.; Foutz, A.; Camacho, M.V.; Raymond, G.J.; Race, B.; Orru, C.D.; Yuan, J.; Shen, P.; Li, B.; et al. Early preclinical detection of prions in the skin of prion-infected animals. Nat. Commun. 2019, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Alibhai, J.; Blanco, R.A.; Barria, M.A.; Piccardo, P.; Caughey, B.; Perry, V.H.; Freeman, T.C.; Manson, J.C. Distribution of Misfolded Prion Protein Seeding Activity Alone Does Not Predict Regions of Neurodegeneration. PLoS Biol. 2016, 14, e1002579. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Del Tredici, K.; Rub, U.; de Vos, R.A.; Jansen Steur, E.N.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Kraus, A.; Anson, K.J.; Raymond, L.D.; Martens, C.; Groveman, B.R.; Dorward, D.W.; Caughey, B. Prion Protein Prolines 102 and 105 and the Surrounding Lysine Cluster Impede Amyloid Formation. J. Biol. Chem. 2015, 290, 21510–21522. [Google Scholar] [CrossRef] [Green Version]
- Groveman, B.R.; Kraus, A.; Raymond, L.D.; Dolan, M.A.; Anson, K.J.; Dorward, D.W.; Caughey, B. Charge neutralization of the central lysine cluster in prion protein (PrP) promotes PrP(Sc)-like folding of recombinant PrP amyloids. J. Biol. Chem. 2015, 290, 1119–1128. [Google Scholar] [CrossRef] [Green Version]
- Han, J.Y.; Jang, H.S.; Green, A.J.E.; Choi, Y.P. RT-QuIC-based detection of alpha-synuclein seeding activity in brains of dementia with Lewy Body patients and of a transgenic mouse model of synucleinopathy. Prion 2020, 14, 88–94. [Google Scholar] [CrossRef] [Green Version]
- Groveman, B.R.; Foliaki, S.T.; Orru, C.D.; Zanusso, G.; Carroll, J.A.; Race, B.; Haigh, C.L. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol. Commun. 2019, 7, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondru, N.; Manne, S.; Greenlee, J.; West Greenlee, H.; Anantharam, V.; Halbur, P.; Kanthasamy, A.; Kanthasamy, A. Integrated Organotypic Slice Cultures and RT-QuIC (OSCAR) Assay: Implications for Translational Discovery in Protein Misfolding Diseases. Sci. Rep. 2017, 7, 43155. [Google Scholar] [CrossRef]
- Abdulrahman, B.A.; Abdelaziz, D.; Thapa, S.; Lu, L.; Jain, S.; Gilch, S.; Proniuk, S.; Zukiwski, A.; Schatzl, H.M. The celecoxib derivatives AR-12 and AR-14 induce autophagy and clear prion-infected cells from prions. Sci. Rep. 2017, 7, 17565. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Wagner, J.; Mitteregger-Kretzschmar, G.; Ryazanov, S.; Leonov, A.; Griesinger, C.; Giese, A. Quantitative Real-Time Quaking-Induced Conversion Allows Monitoring of Disease-Modifying Therapy in the Urine of Prion-Infected Mice. J. Neuropathol. Exp. Neurol. 2015, 74, 924–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manne, S.; Kondru, N.; Hepker, M.; Jin, H.; Anantharam, V.; Lewis, M.; Huang, X.; Kanthasamy, A.; Kanthasamy, A.G. Ultrasensitive Detection of Aggregated alpha-Synuclein in Glial Cells, Human Cerebrospinal Fluid, and Brain Tissue Using the RT-QuIC Assay: New High-Throughput Neuroimmune Biomarker Assay for Parkinsonian Disorders. J. Neuroimmune Pharmacol. 2019, 14, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Hughson, A.G.; Race, B.; Kraus, A.; Sangare, L.R.; Robins, L.; Groveman, B.R.; Saijo, E.; Phillips, K.; Contreras, L.; Dhaliwal, V.; et al. Inactivation of Prions and Amyloid Seeds with Hypochlorous Acid. PLoS Pathog. 2016, 12, e1005914. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Hughson, A.G.; Chesebro, B.; Race, B. Inactivation of chronic wasting disease prions using sodium hypochlorite. PLoS ONE 2019, 14, e0223659. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manca, M.; Kraus, A. Defining the Protein Seeds of Neurodegeneration using Real-Time Quaking-Induced Conversion Assays. Biomolecules 2020, 10, 1233. https://doi.org/10.3390/biom10091233
Manca M, Kraus A. Defining the Protein Seeds of Neurodegeneration using Real-Time Quaking-Induced Conversion Assays. Biomolecules. 2020; 10(9):1233. https://doi.org/10.3390/biom10091233
Chicago/Turabian StyleManca, Matteo, and Allison Kraus. 2020. "Defining the Protein Seeds of Neurodegeneration using Real-Time Quaking-Induced Conversion Assays" Biomolecules 10, no. 9: 1233. https://doi.org/10.3390/biom10091233