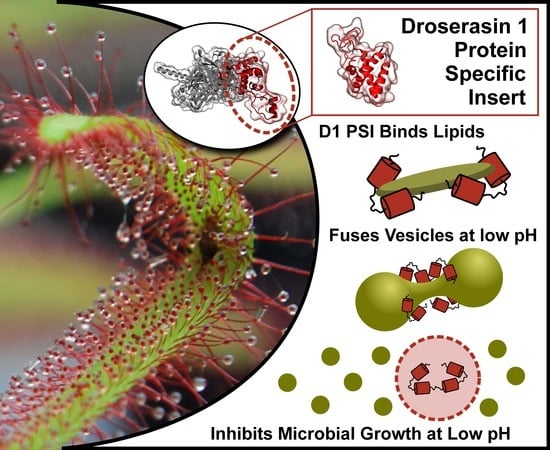
The Droserasin 1 PSI: A Membrane-Interacting Antimicrobial Peptide from the Carnivorous Plant Drosera capensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Alignment and Clustering
2.2. Structure Prediction
2.3. Gene Construction, Expression, and Purification
2.4. Circular Dichroism
2.5. Fluorescence Spectroscopy
2.6. Characterization of Oligomeric State
2.7. Antimicrobial Assay
2.8. Vesicle Fusion Assay
2.9. Lipid Interaction Quantification
2.10. Solid-State NMR
2.11. Molecular Modeling and Analysis
3. Results and Discussion
3.1. D. capensis and Related Plants Contain Several Aspartic Proteases with PSIs
3.2. D. capensis D1 PSI Is Highly Stable
3.3. D1 PSI Is Monomeric in Solution over a Wide pH Range
3.4. D1 PSI Enables Vesicle Fusion at Acidic pH
3.5. D1 PSI Is Able to Interact with Lipids Having Diverse Head Groups
3.6. Solid-State NMR Shows That D1 PSI Is Ordered and Strongly Bound to the Membrane
3.7. Molecular Modeling Suggests Potential Stability of Both Monomeric and Dimeric D1 PSI within Membranes, and Indicates That both Induce Permeability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| D1 | Droserasin 1 |
| DMSO | dimethylsulfoxide |
| DTT | dithiothreitol |
| IPTG | Isopropyl -D-1-thiogalactopyranoside |
| PSI | plant-specific insert |
| MES | 2-(N-morpholino)ethanesulfonic acid |
| POPC | 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine |
| POPE | 1-palmitoyl-2-oleoyl-sn-glycero-3- phosphoethanolamine |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) |
| POPS | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine |
| POPA | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate |
| LUV | large unilamellar vesicles |
| CD | circular dichroism |
| CP | cross-polarization |
| DARR | dipolar-assisted rotational resonance |
| MAS | magic angle spinning |
| MD | molecular dynamics |
| MS | mass spectrometry |
| NMR | nuclear magnetic resonance |
References
- Paniw, M.; Gil-Cabeza, E.; Ojeda, F. Plant carnivory beyond bogs: Reliance on prey feeding Drosophyllum lusitanicum (Drosophyllaceae) in dry Mediterranean heathland habitats. Ann. Bot. 2017, mcw247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokubun, T. Occurrence of myo-inositol and alkyl-substituted polysaccharide in the prey-trapping mucilage of Drosera capensis. Sci. Nat. 2017, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Król, E.; Płachno, B.J.; Adamec, L.; Stolarz, M.; Dziubińska, H.; Trębacz, K. Quite a few reasons for calling carnivores ‘the most wonderful plants in the world’. Ann. Bot. 2011, 109, 47–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palfalvi, G.; Hackl, T.; Terhoeven, N.; Shibata, T.F.; Nishiyama, T.; Ankenbrand, M.; Becker, D.; Förster, F.; Freund, M.; Iosip, A.; et al. Genomes of the Venus flytrap and close relatives unveil the roots of plant carnivory. Curr. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Butts, C.T.; Bierma, J.C.; Martin, R.W. Novel proteases from the genome of the carnivorous plant Drosera capensis: Structural prediction and comparative analysis. Proteins Struct. Funct. Bioinform. 2016, 84, 1517–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butts, C.T.; Zhang, X.; Kelly, J.E.; Roskamp, K.W.; Unhelkar, M.H.; Freites, J.A.; Tahir, S.; Martin, R.W. Sequence comparison, molecular modeling, and network analysis predict structural diversity in cysteine proteases from the Cape sundew, Drosera capensis. Comput. Struct. Biotechnol. J. 2016, 14, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Unhelkar, M.H.; Duong, V.T.; Enendu, K.N.; Kelly, J.E.; Tahir, S.; Butts, C.T.; Martin, R.W. Structure prediction and network analysis of chitinases from the Cape sundew, Drosera capensis. Biochim. Biophys. Acta 2017, 1861, 636–643. [Google Scholar] [CrossRef]
- Duong, V.T.; Unhelkar, M.H.; Kelly, J.E.; Kim, S.H.; Butts, C.T.; Martin, R.W. Network analysis provides insight into active site flexibility in esterase/lipases from the carnivorous plant Drosera capensis. Integr. Biol. 2018, 10, 768–779. [Google Scholar] [CrossRef]
- Bryksa, B.; Bhaumik, P.; Magracheva, E.; De Moura, D.; Kurylowicz, M.; Zdanov, A.; Dutcher, J.; Wlodawer, A.; Yada, R. Structure and mechanism of the saposin-like domain of a plant aspartic protease. J. Biol. Chem. 2011, 286, 28265–28275. [Google Scholar] [CrossRef] [Green Version]
- Bruhn, H. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 2005, 389, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, F.; Palomares-Jerez, M.F.; Daleo, G.; Villalaín, J.; Guevara, M.G. Cholesterol and membrane phospholipid compositions modulate the leakage capacity of the swaposin domain from a potato aspartic protease (StAsp-PSI). Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2011, 1811, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, F.F.; Palomares-Jerez, M.F.; Daleo, G.; Villalaín, J.; Guevara, M.G. Possible mechanism of structural transformations induced by StAsp-PSI in lipid membranes. Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryksa, B.C.; Yada, R.Y. Protein structure insights into the bilayer interactions of the saposin-like domain of Solanum tuberosum aspartic protease. Sci. Rep. 2017, 7, 16911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Hondt, K.; Stack, S.; Gutteridge, S.; Vandekerckhove, J.; Krebbers, E.; Gal, S. Aspartic proteinase genes in the Brassicaceae Arabidopsis thaliana and Brassica napus. Plant Mol. Biol. 1997, 33, 187–192. [Google Scholar] [CrossRef]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef]
- Simoes, I.; Faro, C. Structure and function of plant aspartic proteinases. Eur. J. Biochem. 2004, 271, 2067–2075. [Google Scholar] [CrossRef] [Green Version]
- White, P.C.; Cordeiro, M.C.; Arnold, D.; Kay, J. Processing, activity, and Inhibition of recombinant cyprosin, an aspartic proteinase from cardoon (Cynara cardunculus). J. Biol. Chem. 1999, 274, 16685–16693. [Google Scholar] [CrossRef] [Green Version]
- Duarte, P.; Pissarra, J.; Moore, I. Processing and tracking of a single isoform of the aspartic proteinase cardosin A on the vacuolar pathway. Planta 2008, 227, 1255–1268. [Google Scholar] [CrossRef]
- Pereira, C.; Pereira, S.; Satiat-Jeunemaitre, B.; Pissarra, J. Cardosin A contains two vacuolar sorting signals using different vacuolar routes in tobacco epidermal cells. Plant J. 2013, 76, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Egas, C.; Lavoura, N.; Resende, R.; Brito, R.; Pires, E.; de Lima, M.; Faro, C. The saposin-like domain of the plant aspartic proteinase precursor is a potent inducer of vesicle leakage. J. Biol. Chem. 2000, 275, 38190–38196. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, F.F.; Mendieta, J.R.; Pagano, M.R.; Paggi, R.A.; Daleo, G.R.; Guevara, M.G. The swaposin-like domain of potato aspartic protease (StAsp-PSI) exerts antimicrobial activity on plant and human pathogens. Peptides 2010, 31, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Risør, M.W.; Thomsen, L.R.; Sanggaard, K.W.; Nielsen, T.A.; Thøgersen, I.B.; Lukassen, M.V.; Rossen, L.; Garcia-Ferrer, I.; Guevara, T.; Scavenius, C.; et al. Enzymatic and structural characterization of the major endopeptidase in the Venus flytrap digestion fluid. J. Biol. Chem. 2016, 291, 2271–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, M.; Law, M.; Holt, C.; Stein, J.; Moghe, G.; Hufnagel, D.; Lei, J.; Achawanantakun, R.; Jiao, D.; Lawrence, C.J.; et al. MAKER-P: A Tool-kit for the Rapid Creation, Management, and Quality Control of Plant Genome Annotations. Plant Physiol. 2013, 164, 513–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Malcata, F.X. Proteolysis of ovine caseins by cardosin A, an aspartic acid proteinase from Cynara cardunculus L. Lait 1998, 78, 513–519. [Google Scholar] [CrossRef]
- Silva, S.; Malcata, F. On the activity and specificity of cardosin B, a plant proteinase, on ovine caseins. Food Chem. Toxicol. 1999, 67, 373–378. [Google Scholar] [CrossRef]
- Takahashi, K.; Niwa, H.; Yokota, N.; Kubota, K.; Inoue, H. Widespread tissue expression of nepenthesin-like aspartic protease genes in Arabidopsis thaliana. Plant Physiol. Biochem. 2008, 46, 724–729. [Google Scholar] [CrossRef]
- Kim, D.; Chivian, D.; Baker, D. Protein Structure Prediction and Analysis Using the Robetta Server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [Green Version]
- Raman, S.; Vernon, R.; Thompson, J.; Tyka, M.; Sadreyev, R.; Pei, J.; Kim, D.; Kellogg, E.; DiMaio, F.; Lange, O.; et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 2009, 77, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Faust, G.; Stand, A.; Weuster-Botz, D. IPTG can replace lactose in auto-induction media to enhance protein expression in batch-cultured Escherichia coli. Eng. Life Sci. 2015, 15, 824–829. [Google Scholar] [CrossRef]
- Sivashanmugam, A.; Murray, V.; Cui, C.; Zhang, Y.; Wang, J.; Li, Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009, 18, 936–948. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandana, V.; Karuna, M.S.L.; Vijayalakshmi, P.; Prasad, R.B.N. A simple method to enrich phospholipid content in commercial soybean lecithin. J. Am. Oil Chem. Soc. 2001, 78, 555–556. [Google Scholar] [CrossRef]
- Bird, M.D.; Brey, W.W.; Cross, T.A.; Dixon, I.R.; Griffin, A.; Hannahs, S.T.; Kynoch, J.; Litvak, I.M.; Schiano, J.L.; Toth, J. Commissioning of the 36 T Series-Connected Hybrid Magnet at the NHMFL. IEEE Trans. Appl. Supercond. 2018, 28, 1–6. [Google Scholar] [CrossRef]
- Pines, A.; Gibby, M.G.; Waugh, J.S. Proton-Enhanced Nuclear Induction Spectroscopy. A Method for High Resolution NMR of Dilute Spins in Solids. J. Chem. Phys. 1972, 56, 1776–1777. [Google Scholar] [CrossRef] [Green Version]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar] [CrossRef]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Aksimentiev, A. Script to Exclude Water from Lipid Bilayers. Software File. 2016. Available online: http://www.ks.uiuc.edu/Training/Tutorials/science/membrane/mem-tut-filesr.tar.gz (accessed on 15 July 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3D: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [Green Version]
- Homan, M.D.; Gelman, A. The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res. 2014, 15, 1593–1623. [Google Scholar]
- Stan Development Team. RStan: The R Interface to Stan; R Package Version 2.19.3; 2020. [Google Scholar]
- Rawlings, N.; Waller, M.; Barrett, A.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Athauda, S.; Matsumoto, K.; Rajapakshe, S.; Kuribayashi, M.; Kojima, M.; Kubomura-Yoshida, N.; Iwamatsu, A.; Shibata, C.; Inoue, H. Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: Enzymatic and structural characteristics. Curr. Protein Pept. Sci. 2005, 6, 513–525. [Google Scholar] [CrossRef]
- Buch, F.; Kaman, W.E.; Bikker, F.J.; Yilamujiang, A.; Mithöfe, A. Nepenthesin protease activity indicates digestive fluid dynamics in carnivorous Nepenthes plants. PLoS ONE 2015, 10, e0118853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazorra-Manzano, M.A.; Tanaka, T.; Dee, D.R.; Yada, R.Y. Structure-function characterization of the recombinant aspartic proteinase A1 from Arabidopsis thaliana. Phytochemistry 2010, 71, 515–523. [Google Scholar] [CrossRef]
- Motta, M.; Tatti, M.; Salvioli, R. Autophagic dysfunction in Gaucher disease and its rescue by cathepsin B and D proteases. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Elsevier: Amsterdam, The Netherlands, 2014; pp. 131–146. [Google Scholar] [CrossRef]
- Ahn, V.; Leyko, P.; Alattia, J.; Chen, L.; Prive, G. Crystal structures of saposins A and C. Protein Sci. 2006, 15, 1849–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossmann, M.; Schultz-Heienbrok, R.; Behlke, J.; Remmel, N.; Alings, C.; Sandhoff, K.; Saenger, W.; Maier, T. Crystal structures of human saposins C and D: Implications for lipid recognition and membrane interactions. Structure 2008, 16, 809–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gally, J.; Edelman, G. The effect of temperature on the fluorescence of some aromatic amino acids and proteins. Biochim. Biophys. Acta 1962, 60, 499–509. [Google Scholar] [CrossRef]
- Hennecke, J.; Sillen, A.; Huber-Wunderlich, M.; Engelborghs, Y.; Glockshuber, R. Quenching of tryptophan fluorescence by the active-site disulfide bridge in the DsbA protein from Escherichia coli. Biochemistry 1997, 36, 6391–6400. [Google Scholar] [CrossRef] [PubMed]
- Frauenfeld, J.; Löving, R.; Armache, J.P.; Sonnen, A.F.P.; Guettou, F.; Moberg, P.; Zhu, L.; Jegerschöld, C.; Flayhan, A.; Briggs, J.A.G.; et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods 2016, 13, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.T.H.; Helfinger, L.R.; Bostock, M.J.; Solt, A.; Tan, Y.L.; Nietlispach, D. An adaptable phospholipid membrane mimetic system for solution NMR studies of membrane proteins. J. Am. Chem. Soc. 2017, 139, 14829–14832. [Google Scholar] [CrossRef]
- Flayhan, A.; Mertens, H.D.; Ural-Blimke, Y.; Molledo, M.M.; Svergun, D.I.; Löw, C. Saposin lipid nanoparticles: A highly versatile and modular tool for membrane protein research. Structure 2018, 26, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Ladizhansky, V. Applications of solid-state NMR to membrane proteins. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 1577–1586. [Google Scholar] [CrossRef]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Zilm, K.W. Oral Presentation at the 40th Experimental NMR Conference. Orlando, FL, USA, 1999.
- McDermott, A.; Polenova, T.; Böckmann, A.; Zilm, K.W.; Paulson, E.K.; Martin, R.W.; Montelione, G.T. Partial NMR assignments for uniformly (13C, 15N)-enriched BPTI in the solid state. J. Biomol. NMR 2000, 16, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Igumenova, T.I.; McDermott, A.E.; Zilm, K.W.; Martin, R.W.; Paulson, E.K.; Wand, A.J. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J. Am. Chem. Soc. 2004, 126, 6720–6727. [Google Scholar] [CrossRef]
- Franks, W.T.; Zhou, D.H.; Wylie, B.J.; Money, B.G.; Graesser, D.T.; Frericks, H.L.; Sahota, G.; Rienstra, C.M. Magic-angle spinning solid-state NMR spectroscopy of the β1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J. Am. Chem. Soc. 2005, 127, 12291–12305. [Google Scholar] [CrossRef] [PubMed]
- Castellani, F.; van Rossum, B.; Diehl, A.; Schubert, M.; Rehbein, K.; Oschkinat, H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 2002, 420, 99–102. [Google Scholar] [CrossRef]
- Lange, A.; Becker, S.; Seidel, K.; Giller, K.; Pongs, O.; Baldus, M. A concept for rapid protein-structure determination by solid-state NMR spectroscopy. Angew. Chem. 2005, 44, 2089–2092. [Google Scholar] [CrossRef]
- Sperling, L.J.; Berthold, D.A.; Sasser, T.L.; Jeisy-Scott, V.; Rienstra, C.M. Assignment strategies for large proteins by magic-angle spinning NMR: The 21-kDa disulfide bond forming enzyme DsbA. J. Mol. Biol. 2010, 399, 268–282. [Google Scholar] [CrossRef] [Green Version]
- Manolikas, T.; Herrmann, T.; Meier, B.H. Protein structure determination from 13C spin-diffusion solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 3959–3966. [Google Scholar] [CrossRef]
- Van der Wel, P.C.A. New applications of solid-state NMR in structural biology. Emerg. Top. Life Sci. 2018, 2, 57–67. [Google Scholar]
- Shi, L.; Ahmed, M.A.; Zhang, W.; Whited, G.; Brown, L.S.; Ladizhansky, V. Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump–Structural insights. J. Mol. Biol. 2009, 386, 1078–1093. [Google Scholar] [CrossRef]
- Shahid, S.A.; Bardiaux, B.; Franks, W.T.; Krabben, L.; Habeck, M.; van Rossum, B.J.; Linke, D. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nat. Methods 2012, 9, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Mandala, V.S.; Williams, J.K.; Hong, M. Structure and dynamics of membrane proteins from solid-state NMR. Annu. Rev. Biophys. 2018, 47, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sprague-Piercy, M.A.; Bierma, J.C.; Crosby, M.G.; Carpenter, B.P.; Takahashi, G.R.; Paulino, J.; Hung, I.; Zhang, R.; Kelly, J.E.; Kozlyuk, N.; et al. The Droserasin 1 PSI: A Membrane-Interacting Antimicrobial Peptide from the Carnivorous Plant Drosera capensis. Biomolecules 2020, 10, 1069. https://doi.org/10.3390/biom10071069
Sprague-Piercy MA, Bierma JC, Crosby MG, Carpenter BP, Takahashi GR, Paulino J, Hung I, Zhang R, Kelly JE, Kozlyuk N, et al. The Droserasin 1 PSI: A Membrane-Interacting Antimicrobial Peptide from the Carnivorous Plant Drosera capensis. Biomolecules. 2020; 10(7):1069. https://doi.org/10.3390/biom10071069
Chicago/Turabian StyleSprague-Piercy, Marc A., Jan C. Bierma, Marquise G. Crosby, Brooke P. Carpenter, Gemma R. Takahashi, Joana Paulino, Ivan Hung, Rongfu Zhang, John E. Kelly, Natalia Kozlyuk, and et al. 2020. "The Droserasin 1 PSI: A Membrane-Interacting Antimicrobial Peptide from the Carnivorous Plant Drosera capensis" Biomolecules 10, no. 7: 1069. https://doi.org/10.3390/biom10071069