DMARDs–Gut Microbiota Feedback: Implications in the Response to Therapy
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Disease-Modifying Antirheumatic Drugs (DMARDs) Usage in RA
3.2. Gut Microbiota and csDMARDs’ Metabolism
3.3. Pre- and Post-Treatment Intestinal Dysbiosis with DMARDs in RA
3.3.1. Firmicutes
3.3.2. Bacteroidetes
3.3.3. Proteobacteria
3.3.4. Actinobacteria
3.3.5. Other Bacterial Phyla
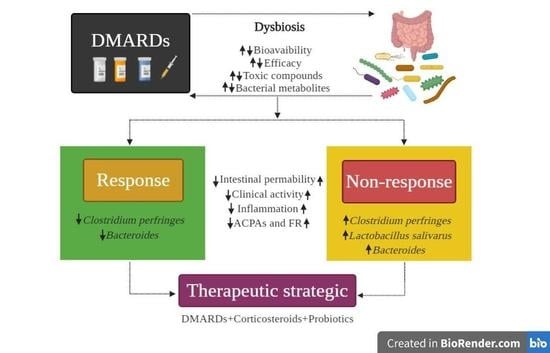
3.4. Gut Microbiota and Its Relation with the Response to Pharmacological Treatment
4. Analysis and Future Perspectives
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Katchamart, W.; Johnson, S.; Lin, H.J.; Phumetthum, V.; Salliot, C.; Bombardier, C. Predictors for remission in rheumatoid arthritis patients: A systematic review. Arthritis Care Res. 2010, 62, 1128–1143. [Google Scholar] [CrossRef] [PubMed]
- Wijbrandts, C.A.; Tak, P.P. Prediction of Response to Targeted Treatment in Rheumatoid Arthritis. Mayo Clin. Proc. 2017, 92, 1129–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinebaum, R.; Neumann, V.C.; Cooke, E.M.; Wright, V. Comparison of faecal florae in patients with rheumatoid arthritis and controls. Rheumatology 1987, 26, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Neumann, V.C.; Shinebaum, R.; Cooke, E.M.; Wright, V. Effects of sulphasalazine on faecal flora in patients with rheumatoid arthritis: A comparison with penicillamine. Br. J. Rheumatol. 1987, 26, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Dearlove, S.M.; Barr, K.; Neumann, V.; Isdale, A.; Bird, H.A.; Gooi, H.C.; Wright, V. The effect of non-steroidal anti-inflammatory drugs on fecal flora and bacterial antibody levels in rheumatoid arthritis. Br. J. Rheumatol. 1992, 31, 443–447. [Google Scholar] [CrossRef]
- Bradley, S.M.; Neumann, V.C.; Barr, K.; Troughton, P.R.; Astbury, C.; Bird, H.A.; Gooi, H.C.; Wright, V. Sequential study of bacterial antibody levels and faecal flora in rheumatoid arthritis patients taking sulphasalazine. Br. J. Rheumatol. 1993, 32, 683–688. [Google Scholar] [CrossRef]
- Eerola, E.; Möttönen, T.; Hannonen, P.; Luukkainen, R.; Kantola, I.; Vuori, K.; Tuominen, J.; Toivanen, P. Intestinal flora in early rheumatoid arthritis. Br. J. Rheumatol. 1994, 33, 1030–1038. [Google Scholar] [CrossRef]
- Peltonen, R.; Nenonen, M.; Helve, T.; Hänninen, O.; Toivanen, P.; Eerola, E. Faecal microbial flora and disease activity in rheumatoid arthritis during a vegan diet. Br. J. Rheumatol. 1997, 36, 64–68. [Google Scholar] [CrossRef] [Green Version]
- Maeda, Y.; Matsushita, M.; Katayama, M.; Yoshimura, M.; Watanabe, A.; Tanaka, E.; Tsuji, S.; Kitatobe, A.; Harada, Y.; Oshima, S.; et al. SAT0079 The analysis of fecal microbiota in rheumatoid arthritis patients compared to healthy volunteers using bacterial RRNA-targeted reverse transcription-quantitative PCR. EULAR Abstracts. Ann. Rheum. Dis. 2012, 71, 496. [Google Scholar] [CrossRef]
- Liu, X.; Zou, Q.; Zeng, B.; Fang, Y.; Wei, H. Analysis of fecal lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 2013, 67, 170–176. [Google Scholar] [CrossRef]
- Maeda, Y.; Matsushita, M.; Yura, A.; Teshigawara, S.; Katayama, M.; Yoshimura, M.; Watanabe, A.; Tanaka, E.; Tsuji, S.; Kitatobe, A.; et al. OP0191 The Fecal Microbiota of Rheumatoid Arthritis Patients Differs from that of Healthy Volunteers and is Considerably Altered by Treatment with Biologics. EULAR Abstracts. Ann. Rheum. Dis. 2013, 72, A117. [Google Scholar] [CrossRef]
- Maeda, Y.; Motooka, D.; Nii, T.; Matsumoto, Y.; Matsushita, M.; Saeki, Y.; Narazaki, M.; Kumanogoh, A.; Nakamura, S.; Takeda, K. AB0139 Investigation of prevotella copri from rheumatoid arthritis patients. EULAR Abstracts. Ann. Rheum. Dis. 2018, 77, 1261. [Google Scholar] [CrossRef]
- Rodrigues, G.S.P.; Cayres, L.C.F.; Gonҫalves, F.P.; Takaoka, N.C.; Lengert, A.H.; Tansini, A.; Brisotti, J.L.; Sasdelli, C.B.G.; Oliveira, G.L.V. Detection of increased relative expression units of Bacteroides and Prevotella, and decreased Clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms 2019, 7, 413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toivanen, P.; Vartiainen, S.; Jalava, J.; Luukkainen, R.; Möttönen, T.; Eerola, E.; Mannienen, R. Intestinal anaerobic bacteria in early rheumatoid arthritis (RA). Arthritis Res. 2002, 4, 5. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Munukka, E.; Korkeamäki, M.; Luukkainen, R.; Toivanen, P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008, 35, 1500–1505. [Google Scholar]
- Gul’neva, M.; Noskov, S.M. Colonic microbial biocenosis in rheumatoid arthritis. Klin. Med. 2011, 89, 45–48. [Google Scholar]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Tap, J.; Abou-Ghantous, J.; Leboime, A.; Nahal, R.S.; Langella, P.; Garchon, H.J.; Chiocchia, G.; Furet, J.P.; Breban, M. Gut microbiota variations correlate with disease activity in spondylarthritis (SpA) and rheumatoid arthritis (RA). ACR Meeting Abstracts. Arthritis Rheumatol. 2014, 662. Available online: https://acrabstracts.org/abstract/gut-microbiota-variations-correlate-withdisease-activity-in-pondyloarthritis-spa-and-rheumatoid-arthritis-ra/ (accessed on 24 January 2020).
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Breban, M.; Tap, J.; Leboime, A.; Said-Nahal, R.; Langella, P.; Chiocchia, G.; Furet, J.P.; Sokol, H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 2017, 76, 1614–1622. [Google Scholar] [CrossRef] [PubMed]
- Piccianti-Diamanti, A.; Penebianco, C.; Salemi, S.; Sorgi, M.L.; Rosa, R.D.; Tropea, A.; Sgrulleti, M.; Salerno, G.; Terracciano, F.; D’Amelio, R.; et al. Analysis of gut microbiota in rheumatoid arthritis patients: Disease-related dysbiosis and modifications induced by etanercept. Int. J. Mol. Sci. 2018, 19, 2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; Kievit, T.; Alfa, M.; Bernstein, C.N.; Domselaar, G.V. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 2018, 6, 221. [Google Scholar] [CrossRef] [PubMed]
- Alpizar-Rodríguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Nayak, R.R.; Alexander, M.; Stapleton-Grey, K.; Ubeda, C.; Scher, U.U.; Turnbaugh, P.J. Perturbation of the human gut microbiome by a non-antibiotic drug contributes to the resolution of autoimmune disease. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Jeong, Y.; Kim, J.W.; You, H.J.; Park, S.J.; Lee, J.; Ju, J.H.; Park, M.S.; Jin, H.; Cho, M.L.; Kwon, B.; et al. Gut microbiota composition and function are altered in patients with early rheumatoid arthritis. J. Clin. Med. 2019, 8, 693. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Mannaa, M.; Kim, Y.; Kim, J.; Kim, G.T.; Seo, Y.S. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes 2019, 10, 748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Chen, Q.; Lin, P.; Xu, R.; He, D.; Ji, W.; Bian, Y.; Shen, Y.; Li, Q.; Liu, C.; et al. Characteristics of gut microbiota in patients with rheumatoid arthrtis in Shangai, China. Front. Cell. Infect. Microbiol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, H.I.; Li, J.R.; Liu, C.C.; Liu, P.Y.; Chen, H.H.; Chen, Y.M.; Lan, J.L.; Chen, D.Y. An association of gut microbiota with different phenotypes in chinese patients with rheumatoid arthritis. J. Clin. Med. 2019, 8, 1770. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Bai, Y.; Zhao, Y.; Lui, Y.; Luo, Y. Intestinal microbiota dynamics in the progression of rheumatoid arthritis. ACR Meeting Abstracts. Arthritis Rheumatol. 2019, 71. Available online: https://acrabstracts.org/abstract/intestinal-microbiota-dynamics-in-theprogression-of-rheumatoid-arthritis/ (accessed on 24 February 2020).
- Hammad, D.B.M.; Hider, S.L.; Liyanapathirana, V.C.; Tonge, D.P. Molecular Characterization of Circulatins Microbiome sigantures in Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2020, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Mena-Vázquez, N.; Ruiz-Limón, P.; Moreno-Indias, I.; Manrique-Arija, S.; Tinahones, F.J.; Fernández-Nebro, A. Expansion of Rare and Harmful Lineages is Associated with Established Rheumatoid Arthritis. J. Clin. Med. 2020, 9, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Muñiz-Pedrogo, D.A.; Chen, J.; Hillmann, B.; Jeraldo, P.; Al-Ghalith, G.; Taneja, V.; Davis, J.M.; Khights, D.; Nelson, H.; Faubion, W.A.; et al. An increased abundance of Clostridiaceae characterizes arthritis in inflammatory bowel disease and rheumatoid arthritis: A cross-sectional study. Inflamm. Bowel Dis. 2019, 25, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Isaac, S.; Artacho, A.; Nayack, R.; Flor, A.; Abramson, S.; Rosenthal, P.; Puchades, L.; Scher, J. OP0119 The pre-treatment gut microbiome predicts early response to rheumatoid therapy. EULAR Abstracts. Ann. Rheum. Dis. 2019, 78, 133–134. [Google Scholar] [CrossRef] [Green Version]
- Cedola, F.; Coras, R.; Sanchez-Lopez, E.; Mateo, L.; Pedersen, A.; Brandy-García, A.; Prior-Español, Á.; Rosental, B.S.; Martínez-Morillo, M.; Guma, M. Choline metabolite is associated with inflammation in arthritis in the elderly. ACR Meeting Abstracts. Arthritis Rheumatol. 2019, 71. Available online: https://acrabstracts.org/abstract/choline-metabolite-is-associated-withi-inflammation-in-arthritis-in-the-elderly/ (accessed on 13 December 2019).
- Isaac, S.; Artacho, A.; Nayack, R.; Abramson, S.B.; Alexander, M.; Koo, I.; Rosenthal, P.; Izmirly, P.; Petterson, A.; Pineda, A.; et al. The pre-treatment gut microbiome predicts early response to metrothexte in rheumatoid arthritis. ACR Meeting Abstracts. Arthritis Rheumatol. 2019, 71. Available online: https://acrabstracts.org/abstract/the-pre-treatment-gut-microbiomepredicts-early-response-to-methotrexate-in-rheumatoid-arthritis/ (accessed on 2 March 2020).
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the japanese population. Ann. Rheum. Dis. 2019, 79, 103–111. [Google Scholar] [CrossRef]
- Gioia, C.; Lucchino, B.; Tarsitano, M.G.; Iannuccelli, C.; Di Franco, M. Dietary Habits and Nutrition in Rheumatoid Arthritis: Can Diet Influence Disease Development and Clinical Manifestations? Nutrients 2020, 12, 1456. [Google Scholar] [CrossRef]
- Rovenský, J.; Svík, K.; Stancíková, M.; Istok, R.; Ebringer, L.; Ferencík, M. Treatment of experimental adjuvant arthritis with the combination of methotrexate and lyophilized Enterococcus faecium enriched with organic selenium. Folia Microbiol. 2002, 47, 573–578. [Google Scholar] [CrossRef]
- Lowe, J.R.; Briggs, A.M.; Whittle, S.; Stephenson, M.D. A systematic review of the effects of probiotic administration in inflammatory arthritis. Complement. Ther. Clin. Pract. 2020, 40, 101207. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Landewé, R.; Bijlsma, J.; Burmester, G.; Chatzidionysiou, K.; Dougados, M.; Nam, J.; Ramiro, S.; Voshaar, M.; van Vollenhoven, R.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann. Rheum. Dis. 2017, 76, 960–977. [Google Scholar] [CrossRef]
- Aviña-Zubieta, J.A.; Galindo-Rodríguez, G.; Newman, S.; Suarez-Almazor, M.E.; Russell, A.S. Long term effectiveness of antimalarial drugs in rheumatics diseases. Ann. Rheum. Dis. 1998, 57, 582–587. [Google Scholar] [CrossRef] [Green Version]
- Cardiel, M.H.; Pons-Estel, B.A.; Sacnun, M.P.; Wojdyla, D.; Saurit, V.; Marcos, J.C.; Pinto, M.R.C.; Cordeiro de Acevedo, A.B.; da Silveria, I.G.; Radominski, S.C.; et al. Treatment of early rheumatoid arthritis in a multinational inception cohort of Latin American Patients: The GLADAR experience. J. Clin. Rheumatol. 2012, 18, 327–335. [Google Scholar] [CrossRef]
- Smolen, J. Treat to target in rheumatology: A historical account on occasion of the 10th aniversary. Rheum. Dis. Clin. N. Am. 2019, 45, 477–485. [Google Scholar] [CrossRef]
- Drosos, A.A.; Pelechas, E.; Voulgari, P.V. Treatment strategies are more important than drugs in the management of rheumatoid arthritis. Clin. Rheumatol. 2020, 39, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Emery, P.; Keystone, E.; Tony, H.P.; Cantagrel, A.; van Vollenhoven, R.; Sanchez, A.; Alecock, E.; Lee, J.; Kremer, J. IL-6 receptor inhibition with tocilizumab improves treatment outcomes in patients with rheumatoid arthritis refractory to anti-tumor necrosis factor biological: Results from a 24-week multicenter randomized placebo-controlled trial. Ann. Rheum. Dis. 2008, 67, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Beaulieu, A.; Rubbert-Roth, A.; Ramos-Remus, C.; Rovensky, J.; Alecock, E.; Woodworth, T.; Alten, R.; OPTION Investigators. Effect of interleukin-6 receptor inhibition with tocilizumab in patients with rheumatoid arthritis (OPTION study): A double-blind, placebo-controlled, randomised trial. Lancet 2008, 371, 987–997. [Google Scholar] [CrossRef]
- Smolen, J.S.; Kay, J.; Doyle, M.K.; Landawé, R.; Matteson, E.L.; Wollenhaupt, J.; Gaylis, N.; Murphy, F.T.; Neal, J.S.; Zhou, Y.; et al. Golimumab in patients with active rheumatoid arthritis after treatment with tumour necrosis factor alpha inhibitors (GO-AFTER study): A multicentre, randomised, double-blind, placebo-controlled, phase III trial. Lancet 2009, 374, 210–221. [Google Scholar] [CrossRef]
- Smolen, J.S.; Landewé, R.; Bijlsma, J.W.J.; Burmester, G.R.; Dougados, M.; Kerschbaumer, A.; McIness, I.B.; Sepriano, A.; van Vollenhoven, R.F.; de Wit, M.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis. 2020, 79, 685–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, J.; Schoels, M.M.; Dörner, T.; Emery, P.; Kvien, T.K.; Smolen, J.S.; Breedveld, F.C. Task Force on the Use of Biosimilars to Treat Rheumatological Diseases. Consensus-based recommendations for the use of biosimilars to treat rheumatological diseases. Ann. Rheum. Dis. 2018, 77, 165–174. [Google Scholar] [CrossRef]
- Haiser, H.J.; Turnbaugh, P.J. Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 2013, 69, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Visser, K.; Van Der Heijde, D. Optimal dosage and route of administration of methotrexate in rheumatoid arthritis: A systematic review of the literature. Ann. Rheum. Dis. 2009, 68, 1094–1099. [Google Scholar] [CrossRef]
- Mack, D.R.; Young, R.; Kaufman, S.S.; Ramey, L.; Vanderhoof, J.A. Methotrexate in patients with crohn’s disease after 6-mercaptopurine. J. Pediatr. 1998, 132, 830–835. [Google Scholar] [CrossRef]
- Widemann, B.C.; Hetherington, M.L.; Murphy, R.F.; Balis, F.M.; Adamson, P.C. Carboxypeptidase-G2 rescue in a patient with high dose methotrexate-induced nephrotoxicity. Cancer 1995, 76, 521–526. [Google Scholar] [CrossRef]
- Buchen, S.; Ngampolo, D.; Melton, R.G.; Hasan, C.; Zoubek, A.; Henze, G.; Bode, U.; Fleischhack, G. Carboxypeptidase G2 rescue in patients with methotrexate intoxication and renal failure. Br. J. Cancer 2005, 92, 480–487. [Google Scholar] [CrossRef] [Green Version]
- Gorostegui, M.; Martínez, E.; Llort, A.; Gros, L.; Dapena, J.L.; Hidalgo, E.; Oliveras, M.; Díaz de Heredia, C.; Bastida, P.; Sánchez de Toledo, J. Carboxipeptidasa G2 (CPDG2) en el rescate de la nefrotoxicidad inducida por metotrexato a altas dosis (MTXHD). An. Pediatr. 2007, 66, 434. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Balis, F.M.; O’Brien, M.M.; Schmiegelow, K.; Pauley, J.L.; Bleyer, A.; Widemann, B.C.; Askenazi, D.; Bergeron, S.; Shirali, A.; et al. Consensus guideline for use of glucarpidase in patients with high-dose methotrexate induced acute kidney injury and delayed methotrexate clearance. Oncologist 2018, 23, 52–61. [Google Scholar] [CrossRef] [Green Version]
- Levy, C.C.; Goldman, P. The enzymatic hydrolysis of methotrexate and folic acid. J. Biol. Chem. 1967, 242, 2933–2938. [Google Scholar]
- Seideman, P.; Beck, O.; Eksborg, S.; Wennberg, M. The pharmacokinetics of methotrexate and its 7-hydroxymetabolite in patients with rheumatoid arthritis. Br. J. Clin. Pharmacol. 1993, 35, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grim, J.; Chladek, J.; Martinkova, J. Pharmacokinetics and pharmacodynamics of methotrexate in non-neoplastic diseases. Clin. Pharmacokinet. 2003, 42, 139–151. [Google Scholar] [CrossRef]
- Treon, S.P.; Chabner, B.A. Concepts in use of high-dose methotrexate therapy. Clin. Chem. 1996, 42, 1322–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Nayak, R.R.; Stapleton-Gray, K.; O’Loughlin, C.; Fischbach, M.; Turnbaugh, P.J. Methotrexate is an antibacterial drug metabolized by human gut bacteria. ACR Meeting Abstracts. Arthritis Rheumatol. 2017, 69. Available online: https://acrabstracts.org/abstract/methotrexate-is-an-antibacterial-drug-metabolized-by-human-gut-bacteria-2/ (accessed on 22 October 2020).
- Zhou, B.; Xia, X.; Wang, P.; Chen, S.; Yu, C.; Huang, R.; Zhang, R.; Wang, Y.; Lu, L.; Yuan, F.; et al. Induction and amelioration of methotrexate-induced gastrointestinal toxicity are related to immune response and gut microbiota. EBioMedicine 2018, 33, 122–133. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Zeng, T.; Wang, Y.; Cui, H.; Wu, J.; Zou, B.; Tao, Z.; Zhang, L.; Garside, G.B.; Tao, S. Dietary restriction increases protective gut bacteria to rescue lethal methotrexate-induced intestinal toxicity. Gut Microbes 2020, 12, 1714401. [Google Scholar] [CrossRef] [Green Version]
- Smolen, J.S.; Landewé, R.; Breedveld, F.C.; Buch, M.; Burmester, G.; Dougados, M.; Emery, P.; Gaujoux-Viala, C.; Gossec, L.; Nam, J.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2013 update. Ann. Rheum. Dis. 2014, 73, 492–509. [Google Scholar] [CrossRef]
- Azadkhan, A.; Truelove, S.; Aronson, J. The disposition and metabolism of sulphasalazine (salicylazosulphapyridine) in man. Br. J. Clin. Pharmacol. 1982, 13, 523–528. [Google Scholar] [CrossRef]
- Deloménie, C.; Fouix, S.; Longuemaux, S.; Brahimi, N.; Picard, B.; Denamur, E.; Dupret, J.M. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: Evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol. 2001, 183, 3417–3427. [Google Scholar] [CrossRef] [Green Version]
- Bishop, J.B.; Witt, K.L.; Gulati, D.K.; MacGregor, J.T. Evaluation of the mutagenicity of the anti-inflammatory drug salicylazosulfapyridine (SASP). Mutagenesis 1990, 5, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Das, K.M.; Eastwood, M.A.; McManus, J.P.; Sircus, W. Adverse reactions during salicylazosulfapyridine therapy and the relation with drug metabolism and acetylator phenotype. N. Engl. J. Med. 1973, 289, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Pullar, T.; Capell, H.A. Variables affecting efficacy and toxicity of sulphasalazine in rheumatoid arthritis. A review. Drugs 1986, 32, 54–57. [Google Scholar] [CrossRef]
- Peppercorn, M.A.; Goldman, P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J. Pharmacol. Exp. Ther. 1972, 181, 555–562. [Google Scholar]
- Rafii, F.; Franklin, W.; Cerniglia, C.E. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl. Environ. Microbiol. 1990, 7, 2146–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafii, F.; Cerniglia, C.E. Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ. Health Perspect. 1995, 103, 17–19. [Google Scholar]
- Fox, R.I. Mechanism of action of leflunomide in rheumatoid arthritis. J. Rheumatol. Suppl. 1998, 53, 20–26. [Google Scholar] [CrossRef]
- Breedveld, F.C.; Dayer, J.M. Leflunomide: Mode of action in the treatment of rheumatoid arthritis. Ann. Rheum. Dis. 2000, 59, 841–849. [Google Scholar] [CrossRef]
- Beynen, A.C.; Van der Molen, A.J.; Geelen, M.J. Inhibition of hepatic cholesterol biosynthesis by chloroquine. Lipids 1981, 16, 472–474. [Google Scholar] [CrossRef]
- Kyburz, D.; Brentano, F.; Gay, S. Mode of action of hydroxychloroquine in RA-evidence of an inhibitory effect on toll-like receptor signaling. Nat. Clin. Pract. Rheumatol. 2006, 2, 458–459. [Google Scholar] [CrossRef]
- Jang, C.H.; Choi, J.H.; Byun, M.S.; Jue, D.M. Chloroquine inhibits production of TNF-α, IL-1β and IL-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes. Rheumatology 2006, 45, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.C.; Mariz, H.A.; Rocha, L.F., Jr.; Oliveira, P.S.; Dantas, A.T.; Duarte, A.L.; Pitta, I.; Galindo, S.L.; Pitta, M.G. Hydroxychloroquine decreases Th17-related cytokines in systemic lupus erythematosus and rheumatoid arthritis patients. Clinics 2013, 68, 766–771. [Google Scholar] [CrossRef]
- Shi, N.; Zahng, S.; Silverman, G.; Li, M.; Cai, J.; Niu, H. Protective effect of hydroxychloroquine on rheumatoid arthritis-associated atherosclerosis. Anim. Models Exp. Med. 2019, 2, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; He, Y.; Tang, J.; Ou, Q.; Lin, J. Alteration of the gut microbiota in tumor necrosis factor-α antagonist-treated collagen-induced arthritis mice. Int. J. Rheum. Dis. 2020, 23, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Mansson, I.; Olhagen, B. Intestinal clostridium perfringens in rheumatoid arthritis and other connective tissue disorders. Studies of fecal flora, serum antitoxin levels and skin hypersensitivity. Acta Rheumatol. Scand. 1966, 12, 167–174. [Google Scholar] [CrossRef]
- Olhagen, B.; Mansson, I. Intestinal clostridium perfringes in rheumatoid arthritis and other collagen diseases. Acta Med. Scand. 1968, 184, 395–402. [Google Scholar] [CrossRef]
- Raymond, N.J.; Henry, J.; Workowski, K.A. Enterococcal arthritis: Case report and review. Clin. Infect. Dis. 1995, 21, 516–522. [Google Scholar] [CrossRef]
- Luo, J.M.; Guo, L.; Chen, H.; Yang, P.F.; Xiong, R.; Peng, Y.; Yang, L. A study of pre-operative presence of micro-organism in affected knee joints of rheumatoid arthritis patients who need total knee arthroplasty. Knee 2017, 24, 409–418. [Google Scholar] [CrossRef]
- de Paiva, C.S.; Jones, D.B.; Stern, M.E.; Bian, F.; Moore, Q.L.; Corbiere, S.; Streckfus, C.F.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; et al. Altered mucosal microbiome diversity and disease severity in Sjögren síndrome. Sci. Rep. 2016, 6, 23561. [Google Scholar] [CrossRef]
- Di Paola, M.; Cavalieri, D.; Albanese, D.; Sordo, M.; Pindo, M.; Donati, C.; Pagnini, I.; Giani, T.; Simonini, G.; Paladini, A.; et al. Alteration of fecal microbiota profiles in juvenile idiopathic arthritis. Associations with HLA-B27 allele and disease status. Front. Microbiol. 2016, 7, 1703. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Halder, C.V.; Faria, A.V.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telesford, K.M.; Yan, W.; Ochoa-Reparaz, J.; Pant, A.; Kircher, C.; Christy, M.A.; Begum-Haque, S.; Kasper, D.L.; Kasper, L.H. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39 (+) Foxp3 (+) T cells and Treg function. Gut Microbes 2015, 6, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erturk-Hasdemir, D.; Kasper, D.L. Finding a needle in a haystack: Bacteroides fragilis polysaccharide A as the archetypical symbiosis factor. Ann. N. Y. Acad. Sci. 2018, 1417, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Troy, E.B.; Kasper, D.L. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front. Biosci. (Landmark Ed.) 2010, 15, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Borer, A.; Weber, G.; Reisenberg, K.; Schlaeffer, F.; Horowitz, J. Septic arthritis due to bacteroides fragilis after pilonidal sinus resection in a patient with rheumatoid arthritis. Clin. Rheumatol. 1997, 16, 632–634. [Google Scholar] [CrossRef] [PubMed]
- Nolla, J.M.; Murillo, O.; Narvaez, J.; Vaquero, C.G.; Lora-Tamayo, J.; Pedrero, S.; Cabo, J.; Ariza, J. Pyogenic arthritis of native joints due to Bacteroides fragilis: Case report and review of the literature. Medicine 2016, 95, e3962. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C. Evidence for immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, B.; Li, S.; Yang, L.; Zhu, D.; Wang, Y.; Wang, H.; Wang, T.; Shi, B.; Gai, Z.; et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid simples from rheumatoid arthrtis or osteoarthritis patients. Sci. Rep. 2018, 8, 14305. [Google Scholar] [CrossRef] [PubMed]
- Maurice, M.M.; Nakamura, H.; Gringhuis, S.; Okamoto, T.; Yoshida, S.; Kullmann, F.; Lechner, S.; van der Voort, E.A.; Leow, A.; Versendaal, J.; et al. Expression of the thioredoxin-thioredoxin reductase system in the inflamed joints of patients with rheumatoid arthritis. Arthritis Rheum. 1999, 42, 2430–2439. [Google Scholar] [CrossRef]
- Kanerud, L.; Scheynius, A.; Nord, C.E.; Hafström, I. Effect of sulphasalazine on gastrointestinal microflora and on mucosa heat schock protein expression in patients with rheumatoid arthritis. Br. J. Rheumatol. 1994, 33, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Kouhsari, E.; Mohammadzadeh, N.; Kashanizadeh, M.G.; Saghafi, M.M.; Hallajzadeh, M.; Fattahi, A.; Ahmadi, A.; Niknejad, F.; Ghafouri, Z.; Asadi, A.; et al. Antimicrobial resistence, prevalence of resistence genes, and molecular characterization in intestinal Bacteroides fragilis group isolates. APMIS 2019, 127, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, S.; Shenoy, P.A.; Chawla, K. Antimicrobial resistance profile and Nim gene among Bacteroides fragilis group isolates in a University Hospital in South India. J. Glob. Infect. Dis. 2019, 11, 59–62. [Google Scholar]
- Bennike, T.B.; Ellingsen, T.; Glerup, H.; Bonderup, O.K.; Carlsen, T.G.; Meyer, M.K.; Bøgsted, M.; Christiansen, G.; Birkelund, S.; Andersen, V.; et al. Proteome analysis of rheumatoid arthritis gut mucosa. J. Proteome Res. 2017, 16, 346–354. [Google Scholar] [CrossRef]
- Gomez, A.; Luckey, D.; Yeoman, C.J.; Marietta, E.V.; Berg Miller, M.E.; Murray, J.A.; White, B.A.; Taneja, V. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS ONE 2012, 7, e36095. [Google Scholar] [CrossRef]
- Asquith, M.; Sternes, P.R.; Costello, M.E.; Karstens, L.; Diamond, S.; Martin, T.M.; Li, Z.; Marshall, M.S.; Spector, T.D.; le Cao, K.A.; et al. HLA Alleles Associated With Risk of Ankylosing Spondylitis and Rheumatoid Arthritis Influence the Gut Microbiome. Arthritis Rheumatol. 2019, 71, 1642–1650. [Google Scholar] [CrossRef]
- Bolin, J.T.; Filman, D.J.; Matthews, D.A.; Hamlin, R.C.; Kraut, J. Cristal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J. Biol. Chem. 1982, 257, 13650–13662. [Google Scholar]
- Filman, D.J.; Bolin, J.T.; Matthews, D.A.; Kraut, J. Cristal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. II. Environmental of bound NADPH and implications for catalysis. J. Biol. Chem. 1982, 257, 13663–13672. [Google Scholar]
- Gargaro, A.R.; Soteriou, A.; Frenkiel, T.A.; Bauer, C.J.; Birdsall, B.; Polshakov, V.I.; Barsukov, I.L.; Roberts, G.C.; Feeney, J. The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate. J. Mol. Biol. 1998, 77, 119–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopytek, S.J.; Dyer, J.C.D.; Knapp, G.S.; Hu, J.C. Resistance to metrothexate due to AcrAB-Dependent export from Escherichia coli. Antimicrob. Agents Chemother. 2000, 44, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäkivuokko, H.; Tiihonen, K.; Tynkkynen, S.; Paulin, L.; Rautonene, N. The effect of age and non-steroidaal anti-inflammatory drugs on human intestinal microbiota composition. Br. J. Nutr. 2010, 103, 227–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, A.P.; Gunasekara, D.B.; Speer, J.; Reed, M.I.; Peña, A.N.; Midkiff, B.R.; Magness, S.T.; Bultman, S.J.; Allbritton, N.L.; Redinbo, M.R. Nonsteroidal anti-inflammatory drug-induced leaky gut modeled using polarized monolayers of primary human intestinal epithelial cells. ACS Infect. Dis. 2018, 4, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, I.; Scarpignato, C.; Holmgren, E.; Olszewski, M.; Rainsford, K.D.; Lanas, A. Mechanisms of damage to the gastrointestinal tract from nonsteroidal anti-inflammatory drugs. Gastroenterology 2018, 154, 500–514. [Google Scholar] [CrossRef] [Green Version]
- Montenegro, L.; Losurdo, G.; Licinio, R.; Zamparella, M.; Giorgio, F.; lerardi, E.; Di Leo, A.; Principi, M. Non-steroidal anti-inflammatory drug induced damage on lower gastro-intestinal tract: Is there an involvement of microbiota? Curr. Drug Saf. 2014, 9, 196–204. [Google Scholar] [CrossRef]
- Otani, K.; Tanigawa, T.; Watanabe, T.; Shimada, S.; Nadatani, Y.; Nagami, Y.; Tanaka, F.; Kamata, N.; Yamagami, H.; Shiba, M.; et al. Microbiota plays a key role in non-steroidal anti-inflammatory drug-induced small intestine damage. Digestion 2017, 95, 22–28. [Google Scholar] [CrossRef]
- Severijnen, A.J.; Kool, J.; Swaak, A.J.G.; Hazenberg, M.P. Intestinal flora of patients with rheumatoid arthritis: Induction of chronic arthritis in rats by cell wall fragments from isolated Eubacterim Aerofaciens strains. Rheumatology 1990, 29, 433–439. [Google Scholar] [CrossRef]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Mirjafary Tafti, Z.S.; Moshiri, A.; Ettehad Marvasti, F.; Tarashi, S.; Sadati Khalili, S.F.; Motanhhary, A.; Fateh, A.; Vaziri, F.; Badi, S.A.; Siadat, S.D. The effect of saturated and unsaturated fatty acids on the production of outer membrane vesicles from Bacteroides fragilis and Bacteroides thetaiotaomicron. Gastroenterol. Hepatol. Bed Bench 2019, 12, 155–162. [Google Scholar]
- Häger, J.; Bang, H.; Hagen, M.; Frech, M.; Träger, P.; Sokolova, M.V.; Steffen, U.; Tascilar, K.; Sarter, K.; Schett, G.; et al. The role of dietary fiber in rheumatoid arthritis patients: A feasibility study. Nutrients 2019, 11, 2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuko, K. A potential Benefit of “Balanced diet” for rheumatoid arthritis. Front. Med. 2018, 5, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chehade, L.; Jaafar, Z.A.; El Masri, D.; Zmerly, H.; Kreidieh, D.; Tannir, H.; Itani, L.; El Ghoch, M. Lifestyle modification in rheumatoid arthritis: Dietary and physical activity recommendations based on evidence. Curr. Rheumatol. Rev. 2019, 15, 209–214. [Google Scholar] [CrossRef]
- Vadell, A.K.E.; Bärebring, L.; Hulander, E.; Gjertsson, I.; Lindgvist, H.M.; Winkvist, A. Anti-inflammatory diet in rheumatoid arthritis (ADIRA)-a randomized, controlled crossover trial indicating effects on disease activity. Am. J. Clin. Nutr. 2020, 11, 1203–1213. [Google Scholar] [CrossRef] [Green Version]
- Badsha, H. Role of diet in influencing rheumatoid arthritis disease activity. Open Rheumatol. J. 2018, 12, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Kumar Singh, A.; Cabral, C.; Kumar, R.; Ganguly, R.; Kumar Rana, H.; Gupta, A.; Lauro, M.R.; Carbone, C.; Reis, F.; Pandey, A.K. Beneficial effects of dietary polyphenols on gut microbiota and strategies to improve delivery efficiency. Nutrients 2019, 11, 2216. [Google Scholar] [CrossRef] [Green Version]
- Zamani, B.; Golkar, H.R.; Farshbaf, S.; Emadi-Baygi, M.; Tajabadi-Ebrahimi, M.; Jafari, P.; Akhavan, R.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 2016, 19, 869–879. [Google Scholar] [CrossRef]
- Vaghef-Mehrabany, E.; Alipour, B.; Homayouni-Rad, A.; Sharif, S.K.; Asghari-Jafarabadi, M.; Zavvari, S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition 2014, 30, 430–435. [Google Scholar] [CrossRef]
- Alipour, B.; Homayouni-Rad, A.; Vaghef-Mehrabany, E.; Sharif, S.K.; Vaghef-Mehrabany, L.; Asgahari-Jafarabadi, M.; Nakhjavani, M.R.; Mohtadi-Nia, J. Effects of Lactobacillus case1 supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: A randomized double-blind clinical trial. Int. J. Rheum. Dis. 2014, 17, 519–527. [Google Scholar]
- Mohammed, A.T.; Khattab, M.; Ahmed, A.M.; Turk, T.; Sakr, N.; Khalil, A.M.; Abdelhalim, M.; Sawaf, B.; Hirayama, K.; Huy, N.T. The therapeutic effect of probiotics on rheumatoid arthritis: A systematic review and meta-analysis of randomized control trials. Clin. Rheumatol. 2017, 36, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Agaeinezhad Rudbane, S.M.; Rahmdel, S.; Abdollahzadeh, S.M.; Zare, M.; Bazrafshan, A.; Mazloomi, S.M. The efficacy of probiotic suppementation in rheumatoid arthritis: A meta-analysis of randomized, controlled trials. Inflammopharmacology 2018, 26, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Guo, R.; Ju, Y.; Wang, Q.; Zhu, J.; Xie, Y.; Zheng, Y.; Li, T.; Liu, Z.; Lu, L.; et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome 2019, 7, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esvaran, M.; Conway, P.L. Lactobacillus fermentum PC1 has the capacity to attenuate joint inflammation in collagen-induced arthritis in DBA/1 mice. Nutrients 2019, 11, 785. [Google Scholar] [CrossRef] [Green Version]
- Shadnoush, M.; Nazemian, V.; Manaheji, H.; Zaringhalam, J. The effect of orally administered probiotics on the behavioral, celular, and molecular aspects of adjuvant-induced arthritis. Basic Clin. Nurosci. 2018, 9, 325–336. [Google Scholar] [CrossRef]
- Marietta, E.V.; Murray, J.A.; Luckey, D.H.; Jeraldo, P.R.; Lamba, A.; Patel, R.; Luthra, H.S.; Mangalam, A.; Taneja, V. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. 2016, 68, 2878–2888. [Google Scholar] [CrossRef] [Green Version]



| References | Country | Phylum | |||
|---|---|---|---|---|---|
| Firmicutes | Bacteroidetes | Proteobacteria | Actinobacteria | ||
| Maeda et al. [20] | Japan | NA | Prevotella copri (↑), Bacteroides (↓) | NA | NA |
| Maeda et al. [12] | Japan | NA | Prevotella (↑) | NA | NA |
| Jeong et al. [26] | Korea | NA | Bacteroidetes (↑) [p = 0.011], Bacteroidia (↑) [p = 0.014], Bacteroidales (↑) [p = 0.014] | NA | Collinsella (↓) [p = 0.004] |
| Liu et al. [10] | China | Lactobacilli (↑) [Lactobacillus salivarus (↑), Lactobacillus iners (↑), Lactobacillus ruminis (↑)] | NA | NA | NA |
| Sun et al. [28] | China | Lactobacillus (↓) [p < 0.001] | Bacteroides (↑) [p = 0.022], Alloprevotella (↓) [p < 0.001], Odoribacter (↓) [p < 0.001] | Escherichia-Shigella (↑) [p = 0.035], Enterobacter (↓) [p < 0.001] | NA |
| Tong et al. [30] | China | Streptococcaceae (↑) [p < 0.001], Lactobacillaceae (↑) [p < 0.001], Enterococcaceae (↑) [p = 0.029], Leuconostacaceae (↑) [p < 0.001] | Bacteroidaceae (↓) [p = 0.011] | NA | NA |
| Gul’neva and Noskov, [16] | Russia | Enterococci (↑), Clostridia (↑), Lactobacteria (↓) | NA | Colibacteria (↑) | NA |
| Toivanen et al. [14] | Finland | NA | Prevotella (↓), Porphyromonas (↓), Bacteroides (↓) [p < 0.001], Bacteroides fragilis (↓) [p < 0.001] | NA | NA |
| Alpizar-Rodríguez et al. [24] | Sweden | Lactobacillaceae (↑) [p = 0.039] | Prevotella spp. (↑) [p = 0.04] | NA | NA |
| Picchianti-Diamanti et al. [22] | Italy | Bacilli (↑) [p = 0.035], Lactobacillaceae (↑) [p = 0.021], Faecalibacterium (↓) [p = 0.012], Faecalibacterium prausnitzii (↓) [p = 0.006] | NA | NA | NA |
| Forbes et al. [23] | Canada | Clostridium III (↑), Faecalicoccus (↑), Streptococcus (↑), Gemmiger (↓), Lachnospira (↓), Roseburia (↓), Sporobacter (↓), Anaerofustis (↓) [p < 0.001], | NA | NA | Actinomyces (↑), Eggerthella (↑), Rhotia (↑) [p < 0.001] |
| Scher et al. [17] | USA | Clostridia (↓), Lachnospiraceae (↓) | Prevotella copri (↑), Bacteroides (↓) | NA | NA |
| References | Country | Phylum | ||||
|---|---|---|---|---|---|---|
| Firmicutes | Bacteroidetes | Proteobacteria | Actinobacteria | Anothers | ||
| Maeda et al. [9] | Japan | Lactobacillus fermetum (↑) [p < 0.01] a, Lactobacillus gasseri (↑) [p < 0.01] a, Lactobacillus ruminis (↑) [p < 0.05] a, Lactobacillus reuteri (↑) [p < 0.05] a, Lactobacillus plantarum (↑) [p < 0.05] a, Enterococcus (↑) [p < 0.01] a | NA | NA | NA | NA |
| Maeda et al. [11] | Japan | Lactobacillus fermetum (↑) a, Lactobacillus gasseri (↑) a, Lactobacillus reuteri (↑) a, Enterococcus (↑) a Clostridium del grupo coccoides (↓) [p = 0.003] a, Subgroup Lactobacillus gasseri (↓) [p = 0.006] a, Subgroup Lactobacillus plantarum (↓) [p = 0.017] a | NA | Enterobacteriaceae (↑) a | Bifidobacterium (↓) [p = 0.027] a | NA |
| Lee et al. [27] | Korea | Clostridium leptum (↑) [p = 0.003] b, Ruthenibacterium lactatiformans (↑) [p = 0.004] b, Anerotruncus colihominis (↑) [p = 0.004] b, Christensenella minuta (↑) [p = 0.024] b, Dialister invisus (↑) [p = 0.030] b, Harryflintia acetispora (↑) [p = 0.045] b | Bacteroidetes (↑) [p = 0.034] b Bacteroides acidifaciens (↑) [p = 0.012] b, Bacteroides faecichinchillae (↑) [p = 0.013] b, Bacteroides clarus (↓) [p = 0.044] b | NA | NA | NA |
| Zhang et al. [33] | China | NA | NA | Haemophilus spp.(↓) c | NA | NA |
| Chiang et al. [29] | China | NA | NA | NA | Collinsella (↑) d | Verrucomicrobiae (↑) [p < 0.05] d, Akkermancia (↑) d |
| Neumann et al. [4] | England | Clostridium perfringes (↓) [p < 0.05] e | NA | Escherichia coli (↓) [p < 0.05] e | NA | NA |
| Bradley et al. [6] | England | Clostridium perfringes (↓) [p < 0.05] e | NA | NA | NA | NA |
| Kanerud et al. [103] | Sweden | Bacillus (↑) [p < 0.05] e | Bacteroides (↓) [p < 0.05] e | Escherichia coli (↓) [p < 0.05] e | NA | NA |
| Breban et al. [21] | France | Prevotellaceae (↓) d, Paraprevotallaceae (↓) d | Proteobacteria (↑) d, Klebsiella (↑) d, Desulfovibrionaceae (↑) d, Succinivibrionaceae (↑) d | Bifidobacterium (↓) d | Tenericutes (↑) d, Synergistetes (↑) d | |
| Picchianti-Diamanti et al. [22] | Italy | Clostridiaceae (↓) [p = 0.05] f | NA | Deltaproteobacteria (↓) [ p = 0.05] f, Enterobacteriales (↓) [p = 0.05] c | NA | Cyanobacteria (↑) [p = 0.016] f, Nostocophycidae (↑) f, Nostocales (↑) [p = 0.031] f |
| Scher et al. [17] | USA | NA | Bacteroides (↑) c | NA | NA | NA |
| Chen et al. [19] | USA | Faecalibacterium (↑) [p < 0.05] c | NA | NA | Eggerthella lenta (↑) d, Collinsella aerofaciens (↑) d | NA |
| Muñiz-Pedrogo et al. [34] | USA | Clostridiaceae (↑) [p = 0.045] d Eubacterium (↑) d | NA | Epsilonproteobacteria (↑) [p = 0.03] d, Campylobacteria, (↑) [p=0.04] d | NA | NA |
| Nayak et al. [25] | USA | NA | Bacteroidetes (↓) c | NA | NA | NA |
| Isaac et al. [37] | USA | Clostridia (↑) [p < 0.05] c | Bacteroidia (↓) [p < 0.05] c | NA | NA | NA |
| Rodrigues et al. [13] | Brazil | Clostridium leptum (↓) [p = 0.005] d | Bacteroides spp. (↑) [p = 0.022] d, Prevotella spp. (↑) [p = 0.023] d | NA | NA | NA |
| Mena-Vázquez et al. [32] | Spain | Enterococcus (↑) [p = 0.008] d, Sedimentibacter (↑) [p = 0.037] d, Dorea formicigenerans (↓) [p = 0.044] d | NA | NA | Collinsella aerofaciens (↑) [p = 0.039] a | NA |
| Author, Year | Country | Time of Study | Therapy Method (n) | Responder (n) | Non-Responder (n) |
|---|---|---|---|---|---|
| Neumann et al. [4] | England | 0–16 weeks | 26 RA (14 SSZ/12 DPA) | NA | NA |
| Bradley et al. [6] | England | 0–16 weeks | 31 RA (SSZ) | ↓Clostridium perfringes in 7 RA patients. ↓IgG antibodies of Clostridium perfringes | ↑Clostridium perfringes in 8 RA patients |
| Kanerud et al. [102] | Sweden | 0–16 weeks | 17 RA (SSZ) | NA | NA |
| Zhang et al. [33] | China | 0–12 weeks | 34 RA (MTX/T2/MTX+T2) | NA | ↑Lactobacillus salivarus |
| Nayak et al. [25] | USA | 0–4 weeks | 23 RA (MTX) | ↓Bacteroidetes in 8 RA patients | ↑Bacteroidetes in 15 RA patients |
| Isaac et al. [37] | USA | 0–16 weeks | 27 RA (MTX) | ↓Bacterial diversity in 11 RA patients | ↑Clostridia and ↓Bacteroidia in 16 RA patients |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaragoza-García, O.; Castro-Alarcón, N.; Pérez-Rubio, G.; Guzmán-Guzmán, I.P. DMARDs–Gut Microbiota Feedback: Implications in the Response to Therapy. Biomolecules 2020, 10, 1479. https://doi.org/10.3390/biom10111479
Zaragoza-García O, Castro-Alarcón N, Pérez-Rubio G, Guzmán-Guzmán IP. DMARDs–Gut Microbiota Feedback: Implications in the Response to Therapy. Biomolecules. 2020; 10(11):1479. https://doi.org/10.3390/biom10111479
Chicago/Turabian StyleZaragoza-García, Oscar, Natividad Castro-Alarcón, Gloria Pérez-Rubio, and Iris Paola Guzmán-Guzmán. 2020. "DMARDs–Gut Microbiota Feedback: Implications in the Response to Therapy" Biomolecules 10, no. 11: 1479. https://doi.org/10.3390/biom10111479