Identification of the NADP+ Structural Binding Site and Coenzyme Effect on the Fused G6PD::6PGL Protein from Giardia lamblia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Purification of the Recombinant G6PD::6PGL Protein
2.2. Effect of the NADP+ Molecule on Protein Stability
2.2.1. Thermal Inactivation Assay
2.2.2. Thermal Stability Assay
2.2.3. Structural Analysis by Circular Dichroism (CD)
2.2.4. Trypsin Digestion
2.2.5. Analysis of the Stability of G6PD::6PGL in the Presence of Guanidine Hydrochloride (Gdn-HCl)
2.3. Determination of the Kd Value of G6PD::6PGL from G. lamblia
2.4. Alignment of the G6PD::6PGL Protein from G. lamblia
2.5. Molecular Docking of the NADP+ Molecule in G6PD::6PGL from G. lamblia
2.6. Molecular Dynamics (MD) Simulations of the NADP+ Molecule in G6PD::6PGL from G. lamblia
3. Results and Discussion
3.1. Effect of the NADP+ Molecule on Protein Stability
3.1.1. Thermal Inactivation Assay
3.1.2. Thermal Stability Assay
3.1.3. Structural Analysis by CD
3.1.4. Trypsin Digestion
3.1.5. Stability of G6PD::6PGL in the Presence of Gdn-HCl
3.2. Determination of the Ligand Dissociation Constant (Kd) of Structural NADP+
3.3. Alignment of the G6PD::6PGL Protein
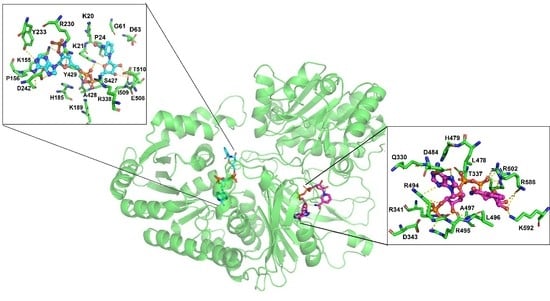
3.4. Molecular Docking of the NADP+ Molecule in G6PD::6PGL from G. lamblia
3.5. MD Simulations of the NADP+ Molecule in G6PD::6PGL from G. lamblia
3.6. Most Populated Cluster Conformation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Plutzer, J.; Ongerth, J.; Karanis, P. Giardia taxonomy, phylogeny and epidemiology: Facts and open questions. Int. J. Hyg. Environ. Health 2010, 213, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Morrison, H.G.; McArthur, A.G.; Gillin, F.D.; Aley, S.B.; Adam, R.D.; Olsen, G.J.; Best, A.A.; Cande, W.Z.; Chen, F.; Cipriano, M.J.; et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 2007, 317, 1921–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einarsson, E.; Ma’ayeh, S.; Svärd, S.G. An up-date on Giardia and giardiasis. Curr. Opin. Microbiol. 2016, 34, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.M.; Stark, D.; Harkness, J.; Ellis, J. Enteric protozoa in the developed world: A public health perspective. Clin. Microbiol. Rev. 2012, 25, 420–449. [Google Scholar] [CrossRef] [Green Version]
- Pierce, K.; Kirkptrick, B. Update on human infections caused by intestinal protozoa. Curr. Opin. Gastroenterol. 2009, 25, 12–17. [Google Scholar] [CrossRef]
- Efstratiou, A.; Ongerth, J.E.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017, 114, 14–22. [Google Scholar] [CrossRef]
- Fink, M.Y.; Singer, S.M. The Intersection of Immune Responses, Microbiota, and Pathogenesis in Giardiasis. Trends Parasitol. 2017, 33, 901–913. [Google Scholar] [CrossRef]
- Bartelt, L.A.; Platts-Mills, J.A. Giardia: A pathogen or commensal for children in high-prevalence settings? Curr. Opin. Infect. Dis. 2015, 29, 502–507. [Google Scholar] [CrossRef]
- Edlind, T.D.; Hang, T.L.; Chakraborty, P.R. Activity of the anthelmintic benzimidazoles against Giardia lamblia in vitro. J. Infect. Dis. 1990, 162, 1408–1411. [Google Scholar] [CrossRef]
- Hilario, E.; Gogarten, J.P. The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J. Mol. Evol. 1998, 46, 703–715. [Google Scholar] [CrossRef]
- Duffieux, F.; Van Roy, J.; Michels, P.A.; Opperdoes, F.R. Molecular characterization of the first two enzymes of the pentose-phosphate pathway of Trypanosoma brucei. Glucose-6-phosphate dehydrogenase and 6-phosphogluconolactonase. J. Biol. Chem. 2000, 275, 27559–27565. [Google Scholar] [PubMed] [Green Version]
- Cosgrove, M.S.; Gover, S.; Naylor, C.E.; Vandeputte-Rutten, L.; Adams, M.J.; Levy, H.R. An examination of the role of asp-177 in the His-Asp catalytic dyad of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase: X-ray structure and pH dependence of kinetic parameters of the D177N mutant enzyme. Biochemistry 2001, 39, 15002–15011. [Google Scholar] [CrossRef] [PubMed]
- Au, S.W.; Naylor, C.E.; Gover, S.; Vandeputte-Rutten, L.; Scopes, D.A.; Mason, P.J.; Luzzatto, L.; Lam, V.M.; Adams, M.J. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 826–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotaka, M.; Gover, S.; Vandeputte-Rutten, L.; Au, S.W.; Lam, V.M.S.; Adams, M.J. Structural studies of glucose-6-phosphate and NADP(+) binding to human glucose-6-phosphate dehydrogenase. Acta Crystallogr. 2005, 61, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Au, S.W.; Gover, S.; Lam, V.M.; Adams, M.J. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure 2000, 8, 293–303. [Google Scholar] [CrossRef] [Green Version]
- Fiorelli, G.; Martinez di Montemuros, F.; Cappellini, M.D. Chronic non-spherocytic haemolytic disorders associated with glucose-6-phosphate dehydrogenase variants. Best Pract. Res. Clin. Haematol. 2000, 13, 39–55. [Google Scholar] [CrossRef]
- Vulliamy, T.J.; D’Urso, M.; Battistuzzi, G.; Estrada, M.; Foulkes, N.S.; Martini, G.; Calabrof, V.; Poggi, V.; Giordano, R.; Town, M.; et al. Diverse point mutations in the human glucose-6-phosphate dehydrogenase gene cause enzyme deficiency and mild or severe hemolytic anemia. Proc. Natl. Acad. Sci. USA 1998, 85, 5171–5175. [Google Scholar] [CrossRef] [Green Version]
- Stover, N.A.; Dixon, T.A.; Cavalcanti, A.R. Multiple Independent Fusions of Glucose-6-Phosphate Dehydrogenase with Enzymes in the Pentose Phosphate Pathway. PLoS ONE 2011, 6, e22269. [Google Scholar] [CrossRef] [Green Version]
- Morales-Luna, L.; Serrano-Posada, H.; González-Valdez, A.; Ortega-Cuellar, D.; Vanoye-Carlo, A.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Rufino-González, Y.; Castillo-Rodríguez, R.A.; Pérez de la Cruz, V.; et al. Biochemical Characterization and Structural Modeling of Fused Glucose-6-Phosphate Dehydrogenase-Phosphogluconolactonase from Giardia lamblia. Int. J. Mol. Sci. 2018, 19, 2518. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Gómez-Manzo, S.; Terrón-Hernández, J.; De la Mora-De la Mora, I.; González-Valdez, A.; Marcial-Quino, J.; García-Torres, I.; Vanoye-Carlo, A.; López-Velázquez, G.; Hernández-Alcántara, G.; Oria-Hernández, J.; et al. The Stability of G6PD Is Affected by Mutations with Different Clinical Phenotypes. Int. J. Mol. Sci. 2014, 15, 21179–21201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Enríquez-Flores, S.; De la Mora-De la Mora, I.; González-Valdez, A.; García-Torres, I.; Martínez-Rosas, V.; Sierra-Palacios, E.; Lazcano-Pérez, F.; et al. Mutations of Glucose-6-Phosphate Dehydrogenase Durham, Santa-Maria and A+ Variants Are Associated with Loss Functional and Structural Stability of the Protein. Int. J. Mol. Sci. 2015, 16, 28657–28668. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Serrano-Posada, H.; González-Valdez, A.; Martínez-Rosas, V.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Castillo-Rodríguez, R.A.; Cuevas-Cruz, M.; et al. Functional and biochemical characterization of three recombinant human Glucose-6-Phosphate Dehydrogenase mutants: Zacatecas, Vanua-Lava and Viangchan. Int. J. Mol. Sci. 2016, 17, 787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonyuen, U.; Chamchoy, K.; Swangsri, T.; Junkree, T.; Day, M.; White, N.; Imwong, M. A trade-off between catalytic activity and protein stability determines the clinical manifestations of glucose-6-phosphate dehydrogenase (G6PD) deficiency. Int. J. Biol. Macromol. 2017, 104, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Nava, E.J.; Ortega-Cuellar, D.; Serrano-Posada, H.; González-Valdez, A.; Vanoye-Carlo, A.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Hernández-Pineda, J.; Rodríguez-Bustamante, E.; Arreguin-Espinosa, R.; et al. Biochemical Analysis of Two Single Mutants that Give Rise to a Polymorphic G6PD A-Double Mutant. Int. J. Mol. Sci. 2017, 18, 2244. [Google Scholar] [CrossRef] [Green Version]
- Cortés-Morales, Y.Y.; Vanoye-Carlo, A.; Castillo-Rodríguez, R.A.; Serrano-Posada, H.; González-Valdez, A.; Ortega-Cuellar, D.; Hernández-Ochoa, B.; Moreno-Vargas, L.M.; Prada-Gracia, D.; Sierra-Palacios, E.; et al. Cloning and biochemical characterization of three glucose 6 phosphate dehydrogenase mutants presents in the Mexican population. Int. J. Biol. Macromol. 2018, 119, 926–936. [Google Scholar] [CrossRef]
- Wang, X.T.; Chan, T.F.; Lam, V.; Engel, P. What is the role of the second “structural” NADP-binding site in human glucose-6-phosphate dehydrogenase? Protein Sci. 2008, 17, 1403–1411. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 16, 2525–2534. [Google Scholar] [CrossRef] [Green Version]
- Weiner, S.J.; Kollman, P.A.; Case, D.A.; Singh, U.C.; Ghio, C.; Alagona, G.; Profeta, S., Jr.; Weiner, P.J. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; DeLano Scientific: Palo Alto, CA, USA, 2002.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmberg, N.; Ryde, U.; Bülow, L. Redesign of the coenzyme specificity in L-lactate dehydrogenase from bacillus stearothermophilus using site-directed mutagenesis and media engineering. Protein Eng. 1999, 12, 851–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impery, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Choi, M.Y.; Au, S.W.; Au, D.M.; Lam, V.M.S.; Engel, P.C. Purification and detailed study of two clinically different human glucose 6-phosphate dehydrogenase variants, G6PD (Plymouth) and G6PD (Mahidol): Evidence for defective protein folding as the basis of disease. Mol. Genet. Metab. 2008, 93, 44–53. [Google Scholar] [CrossRef]
- Beutler, E.; Collins, Z. Hybridization of Glucose-6-Phosphate Dehydrogenase from Rat and Human Erythrocytes. Science 1965, 150, 1306–1307. [Google Scholar] [CrossRef]
- Yoshida, A.; Stamatoyannopoulos, G.; Motulsky, A.G. Negro variant of glucose-6-phosphate dehydrogenase deficiency (A−) in man. Science 1967, 155, 97–99. [Google Scholar] [CrossRef]
- Gómez-Gallego, F.; Garrido-Pertierra, A.; Mason, P.J.; Bautista, J.M. Unproductive folding of the human G6PD-deficient variant A−. FASEB J. 1996, 10, 153–158. [Google Scholar] [CrossRef]
- Wang, X.T.; Lam, V.M.; Engel, P.C. Marked decrease in specific activity contributes to disease phenotype in two human glucose 6-phosphate dehydrogenase mutants, G6PD (Union) and G6PD (Andalus). Hum. Mutat. 2005, 26, 284. [Google Scholar] [CrossRef] [PubMed]









© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Luna, L.; González-Valdez, A.; Sixto-López, Y.; Correa-Basurto, J.; Hernández-Ochoa, B.; Cárdenas-Rodríguez, N.; Castillo-Rodríguez, R.A.; Ortega-Cuellar, D.; Arreguin-Espinosa, R.; Pérez de la Cruz, V.; et al. Identification of the NADP+ Structural Binding Site and Coenzyme Effect on the Fused G6PD::6PGL Protein from Giardia lamblia. Biomolecules 2020, 10, 46. https://doi.org/10.3390/biom10010046
Morales-Luna L, González-Valdez A, Sixto-López Y, Correa-Basurto J, Hernández-Ochoa B, Cárdenas-Rodríguez N, Castillo-Rodríguez RA, Ortega-Cuellar D, Arreguin-Espinosa R, Pérez de la Cruz V, et al. Identification of the NADP+ Structural Binding Site and Coenzyme Effect on the Fused G6PD::6PGL Protein from Giardia lamblia. Biomolecules. 2020; 10(1):46. https://doi.org/10.3390/biom10010046
Chicago/Turabian StyleMorales-Luna, Laura, Abigail González-Valdez, Yudibeth Sixto-López, José Correa-Basurto, Beatriz Hernández-Ochoa, Noemí Cárdenas-Rodríguez, Rosa Angélica Castillo-Rodríguez, Daniel Ortega-Cuellar, Roberto Arreguin-Espinosa, Verónica Pérez de la Cruz, and et al. 2020. "Identification of the NADP+ Structural Binding Site and Coenzyme Effect on the Fused G6PD::6PGL Protein from Giardia lamblia" Biomolecules 10, no. 1: 46. https://doi.org/10.3390/biom10010046