1. Introduction
Due to the growth of advanced medical imaging modalities, it is very difficult to analyze the medical images manually. For this reason, an advanced and efficient computer-aided system is needed to diagnose the diseases. This will help the hematologist to begin the treatment at the right time and increase the patient’s survival rate. Leukemia is a cancer of blood-forming tissues that affects the bone marrow. Leukemia is caused by the proliferation of abnormal white blood cells in the body. Leukemia is mostly affected by people living in developed countries and children aged 14 or under. As per the National Cancer Institute (NCI) statistics, in the United States, it is expected that there will be 62,130 persons as new cases for cancer treatment and 245,000 cases that are fatal or very serious [
1]. In India, leukemia stands at ninth position among diseases (tumors) among children [
2,
3]. Leukemia is identified into two broad categories such as acute and chronic. Acute forms of leukemia occur when the number of immature blood cells increases, and it is the most common type of leukemia in children. Segmenting an image of a nucleus is one of the major challenging tasks in leukemia diagnosis. Recently, soft computing plays an important role in many research areas such as medical image processing, pattern recognition, big data analytics, Internet of Things (IoT) analysis, bioinformatics, and so on.
The rough set theory [
4] was proposed by Pawlak in 1982. This concept is an extension of set theory for the study of intelligent systems characterized by insufficient and incomplete information. This classical rough set theory is based on equivalence relations, but it can also be extended to covering based rough sets [
5,
6,
7]. In 1999, Molodtsov [
8] proposed the concept of a soft set, which can be seen as a new mathematical approach to vagueness. The absence of any restrictions on the approximate description in soft set theory makes this theory very versatile and easily applicable in practice. Maji et al. [
9] improved Molodtsov’s idea by introducing several operations in soft set theory. In [
10], the researcher investigated a soft covering-based rough set as a new kind of soft rough set. This method is a combination of a covering soft set and rough set. In [
11], a covering-based rough k-means clustering approach is applied to segment the leukemia nucleus. The advantage of covering-based subsets is that they generate upper and lower approximations by using the covering feature, which brings about more roughness. Since different clusters give rise to different results, determination of the number of clusters is a difficult task in clustering-based segmentation. To overcome this limitation, the hybrid histogram-based soft covering rough k-means clustering algorithm (HSCRKM) is introduced to segment the image of the leukemia nucleus. In this algorithm, the peak values of the histogram of an image are identified and the number of clusters is initialized. This will avoid the random initialization of a number of clusters. Here, soft covering approximation space is also included. The term ‘covering soft set’ is more accurate than ‘soft rough set.’ It also combines the strengths of covering soft set theory and the rough k-means clustering algorithm to effectively segment the image of the nucleus. Soft covering rough approximation is utilized to find the lower and upper approximation values. The performance of the HSCRKM algorithm is evaluated using existing algorithms such as k-means clustering, fuzzy c-means clustering, and particle swarm optimization (PSO)-based clustering. Different types of features such as GLCM-0, GLCM-45, GLCM-90, GLCM-135, and shape color-based features are extracted from the segmented leukemia nucleus image. Nowadays, a lot of machine learning algorithms are applied to predict the degree of sickness. The state-of-art machine learning prediction algorithms such as neural networks (NN) [
12], logistic regression (LR) [
13], support vector machine (SVM) [
14], naive Bayes (NB) [
15], k-nearest neighborhood (KNN) [
13], decision tree (DT) [
13], and random forest (RF) [
16] are applied to classify the cancerous and non-cancerous leukemia cells. The empirical results show that logistic regression and neural network efficiently predict the blast and non-blast cells when compared with other prediction algorithms.
The main objective of this research work is to develop a diagnostic approach for the identification of acute lymphoblastic leukemia blast cells using image processing and computational intelligence techniques. In experimental analysis, relevant image processing and computational intelligence techniques are applied in order to select the most suitable approach for the delineation of acute lymphoblastic leukemia cells. The following objectives have been formulated in order to predict leukemia: to apply computational intelligence-based algorithms for the segmentation of acute lymphoblastic leukemia blast cells in images and to apply machine learning algorithms to evaluate the performance of the proposed method.
The contribution of this study is summarized as follows. To find the number of clusters using the peak value of a histogram image and compute the lower and upper approximation values based on the soft covering approximation space, three clustering methods—k-means, FCM, and PSO-based clustering—are preferred for segmentation comparison. Through these methods, different kinds of features are extracted, and the efficiency of the proposed algorithm is assessed using machine learning prediction algorithms. The HSCRKM achieves the successful results i.e., above 80% when compared with the existing clustering algorithms. Therefore, it can be concluded that the HSCRKM clustering algorithm works effectively with the other clustering algorithms.
In the clustering algorithm, defining the number of clusters is a very difficult task. To overcome this limitation, the proposed algorithm identifies the peak values of the histogram of an image and initializes the number of clusters. This is one of the advantages of our proposed method, which avoids the random initialization of a number of clusters. The next advantage of the HSCRKM algorithm is that it combines the strengths of covering soft set theory and the rough k-means clustering algorithm to effectively segment the image of the nucleus. Based on a literature review, the term ‘covering soft set’ is more accurate than ‘soft rough set’, since it gives a better result than the soft rough set for several applications. In covering rough sets, the lower and upper approximation values are computed based on the soft covering approximation space.
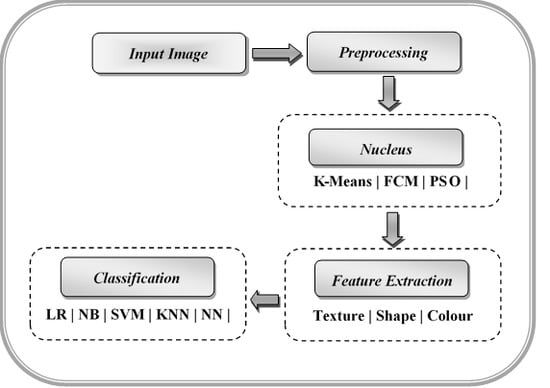
Morphologically, a lymphoblast consists of a massive nucleus of irregular shape and size. In blood sample images, it is difficult to identify the cytoplasm, because it appears rarely and even if it does, it looks intensely colored. The nucleus and cytoplasm of lymphoblast cells reflect the morphological and functional changes. Feature extraction plays a main role in the assessment of leukemia in blood samples. After segmenting the nucleus using the proposed HSCRKM algorithm, salient features are extracted. It reduces the amount of data space and the working time of an image. In this research, different kinds of features are extracted such as gray level co-occurrence matrix (GLCM), color, and shape-based features. These were measured from every channel of the segmented nucleus image. The efficiency of the proposed algorithm is assessed using machine learning prediction algorithms. The performance of the segmentation algorithms was analyzed in the light of different machine learning (ML) prediction methods. With respect to HSCRKM clustering algorithms, most of the ML methods (except naive Bayes) achieved greater than 80% prediction accuracy compared with the existing clustering algorithms, viz., k-means clustering, fuzzy c-means clustering, and rough k-means clustering. It is inferred that the proposed clustering algorithms are more effective in segmenting the nucleus image. Due to the effective segmentation process, the extracted features have increased the prediction accuracy. To evaluate the experimental results, we have empirically set the best accuracy to be greater than 80%. The outline of the proposed system is shown in
Figure 1.
The rest of the research report is organized as follows.
Section 2 reviews the related literature on clustering-based segmentation algorithms.
Section 3 describes the methods of the proposed algorithm and its results. The empirical results are discussed in
Section 4.
Section 5 states the conclusion and indicates the future direction of this research.
2. Related Literature
In recent years, a lot of clustering algorithms have been developed for segmenting medical images.
Petal [
17] applied k-means clustering for segmentation and the Zack algorithm for clustered white blood cells (WBCs). The features—namely, the mean, standard deviation, area, elongation, perimeter, color etc.—are extracted, and support vector machine (SVM) was used to classify the cells. The proposed algorithm effectively segmented the WBCs, which produced 93.57% accuracy. For this experiment, 27 images from the Acute Lymphoblastic Leukemia Image Database (ALL-IDB) were utilized.
Two bare-bones particle swarm optimization (BBPSO) algorithms with and without subswarms were introduced by Srisukkham et al. in 2017 [
18] to diagnosis the leukemia cells. A stimulating discriminant measure (SDM)-based clustering algorithm that combined with the genetic algorithm (GA) was employed to segment the nucleus, cytoplasm, and background regions. The relevant features were extracted; then, various feature selection methods such as particle swarm optimization (PSO), cuckoo search (CS), and dragonfly algorithm (DA) were applied to select the optimal features and reduce the dimensions. An average geometric mean was computed with different sizes of training and test samples to evaluate the performance of the proposed methods. The BBPSO and binary BBPSO algorithms produced 91% to 96% of the geometric mean value.
Su [
19] developed two stages of segmentation process using k-means clustering and HMRF (hidden Markov random field), which are used to group the six different types of AML cells from the bone marrow images. The segmentation algorithm achieved an accuracy of 96% to 98% (average) when compared with other existing segmentation methods.
In [
20], k-means and fuzzy c-means clustering algorithms were applied to segment the brain tumor images. Various feature reduction algorithms, namely probabilistic principal component analysis (PPCA), expectation maximization-based principal component analysis (EM-PCA), the generalized Hebbian algorithm (GHA), and adaptive principal component extraction (APEX) were employed to reduce the dimensions of the feature set. The produced coefficient of variance (CV) values for k-means and Fuzzy C-mean (FCM) are 0.4582 and 0.1224, respectively.
In [
21], potential field segmentation was employed to segment the MRI brain tumor images. This method achieved the standard deviation of 0.283, the average value of 0.517, and the median values of 0.644. From the experimental results, it was observed that ensemble methods generated better segmentations.
Küçükkülahlı [
22] and Namburu [
23] identified the number of cluster values in the clustering algorithm using the peak value of the histogram of an image. In [
22], the automatic segmentation method using the histogram-based k-means clustering algorithm was developed. In [
23], the soft fuzzy rough c-means clustering algorithm (SFRCM) was used to segment the MRI brain tumor images. The proposed SRFCM algorithm achieved a better Jaccard coefficient value of 0.97 for without noise and 0.79 for with 9% Gaussian noise when compared with the existing clustering algorithms namely, k-means, rough k-means (RKM), rough fuzzy c-means (RFCM), and generalized rough c-means (GFCM).
Ali [
24] introduced a new clustering algorithm based on neutrosophic orthogonal matrices (CANOM) to segment the dental X-ray images. The experimental results show that the CANOM simplified silhouette width criterion (SSWC) index is 0.941 and the FCM is 0.02. CANOM is also better than Otsu and eSFCM with the values being 0.657 and 0.647, respectively. The value of CANOM is 47 times larger than that of FCM and 1.43 times larger than those of Otsu and eSFCM.
In [
25], the unsupervised fuzzy c-means (FCM) clustering technique was employed for prostate cancer MRI images. The derived average dice similarity, Jaccard index, sensitivity, specificity, mean absolute difference, and Hausdorff distance is 88.68, 81.26, 90.71, 88.09, 88.09, 3.5, and 4.1 respectively.
In [
26], the proposed multi-Otsu thresholding-based segmentation method can successfully segmented the CT image stacks. In addition, it sows the distribution characteristics of different components in three dimensions.
In [
27], the enhanced adaptive fuzzy k-means (AFKM) algorithm was used to detect the three regions such as white matter (WM), gray matter (GM), and cerebrospinal fluid spaces (CSF) in the brain images. AFKM performed better than FCM, which produced a minimum mean square error (MSE) value of 2.2441.
In [
28], the clustering method intuitionistic fuzzy c-means (IFCM) was applied for medical image segmentation. It is observed from the experimental results that the proposed method outperformed other algorithms that achieved the average quantitative index 0.95. The chronic wound region was detected using fuzzy spectral clustering in [
29]. The proposed method produced 91.5% segmentation accuracy, an 86.7% Dice index, a Jaccard score of 79.0%, 87.3% sensitivity, and 95.7% specificity.
In [
30], the convolutional neural networks (CNN) approach is applied to identify the subtypes of leukemia. It is observed from the experimental results that the CNN model achieves 88.25% and 81.74% accuracy for leukemia and healthy cells, respectively. From the literature review, it is inferred that the clustering-based algorithms were applied to segment the tumor region. A brief review of the literature on various clustering methods in image segmentation and their performances appears in
Table 1.
5. Conclusions and Future Work
Clustering is an unsupervised classification method that is widely employed for image segmentation. Throughout the present research, a hybrid histogram-based soft covering rough k-means clustering algorithm is proposed to segment the image of the leukemia nucleus. In this method, the histogram is used to initialize the number of clusters. The main advantage of this method is that it applies the soft covering rough approximation instead of rough approximation. It is a new kind of soft rough set that efficiently deals with uncertainties. The results are interpreted in the following two ways. (1) The efficiency of the proposed technique is compared with the popular and frequently used clustering algorithms such as k-means clustering, FCM, and PSO-based clustering. (2) The state-of-the-art prediction techniques in machine learning (ML) were compared using evolution metrics.
From the experimental results, it is inferred that the HSCRKM clustering algorithm and all of the ML methods (except for naive Bayes) achieve above 80% prediction accuracy. It is also noted that logistic regression and neural network provide on average above 90% accuracy, which performs better than other prediction methods. The limitation of this method is that when we go for multiple color images such as satellite images, agricultural images, photographs etc., the number of peak values in the histogram is increased, and consequently the processing time is also increased. This method is more suitable for the segmentation of medical images and the extraction of specific portions with high clarity (for deep study). In the future, bio-inspired algorithms could be used to optimize the number of clusters.