Cadmium Sulfide Quantum Dots, Mitochondrial Function and Environmental Stress: A Mechanistic Reconstruction through In Vivo Cellular Approaches in Saccharomyces cerevisiae
Abstract
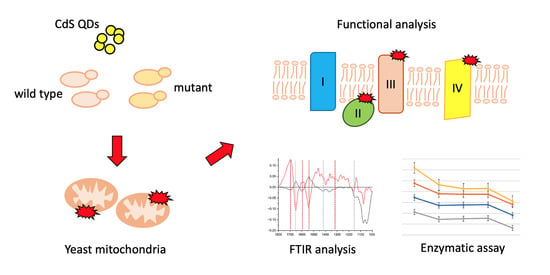
:1. Introduction
2. Methods
2.1. Experimental Setup
2.2. Cadmium Sulfide Quantum Dots Characterization
2.3. Mitochondrial Extraction
2.4. Fourier-Transform Infrared Spectroscopy (FTIR) Measurements
2.5. Enzymatic Assays
3. Results and Discussion
3.1. FTIR Analyses

3.2. Colorimetric Assays and Comparison with FTIR

4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altmann, K.; Dürr, M.; Westermann, B. Saccharomyces cerevisiae as a Model Organism to Study Mitochondrial Biology. Mitochondria 2007, 372, 81–90. [Google Scholar] [CrossRef]
- Betlem, K.; Hoksbergen, S.; Mansouri, N.; Down, M.; Losada-Pérez, P.; Eersels, K.; van Grinsven, B.; Cleij, T.; Kelly, P.; Sawtell, D.; et al. Real-time analysis of microbial growth by means of the Heat-Transfer Method (HTM) using Saccharomyces cerevisiae as model organism. Phys. Med. 2018, 6, 1–8. [Google Scholar] [CrossRef]
- Guaragnella, N.; Coyne, L.P.; Chen, X.J.; Giannattasio, S. Mitochondria–cytosol–nucleus crosstalk: Learning from Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy088. [Google Scholar] [CrossRef] [PubMed]
- Giaver, G.; Nislow, C. The yeast deletion collection: A decade of functional genomics. Genetics 2014, 197, 451–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmiroli, M.; Pagano, L.; Pasquali, F.; Zappettini, A.; Tosato, V.; Bruschi, C.V.; Marmiroli, N. A genome-wide nanotoxicology screen of Saccharomyces cerevisiae mutants reveals the basis for cadmium sulphide quantum dot tolerance and sensitivity. Nanotoxicology 2016, 10, 84–93. [Google Scholar] [CrossRef]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef]
- Pasquali, F.; Agrimonti, C.; Pagano, L.; Zappettini, A.; Villani, M.; Marmiroli, M.; White, J.C.; Marmiroli, N. Nucleo-mitochondrial interaction of yeast in response to cadmium sulfide quantum dot exposure. J. Hazard. Mater. 2017, 324, 744–752. [Google Scholar] [CrossRef]
- Iavicoli, I.; Leso, V.; Fontana, L.; Calabrese, E.J. Nanoparticle Exposure and Hormetic Dose–Responses: An Update. Int. J. Mol. Sci. 2018, 19, 805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paesano, L.; Marmiroli, M.; Bianchi, M.G.; White, J.C.; Bussolati, O.; Zappettini, A.; Villani, M.; Marmiroli, N. Differences in toxicity, mitochondrial function and miRNome in human cells exposed in vitro to Cd as CdS quantum dots or ionic Cd. J. Hazard. Mater. 2020, 393, 122430. [Google Scholar] [CrossRef] [PubMed]
- Bruchez, M., Jr.; Moronne, M.; Gin, P.; Weiss, S.; Alivisatos, A.P. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 1998, 281, 2013–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, H.; Careem, M.; Arof, A. Quantum dot-sensitized solar cells—Perspective and recent developments: A review of Cd chalcogenide quantum dots as sensitizers. Renew. Sustain. Energy Rev. 2013, 22, 148–167. [Google Scholar] [CrossRef]
- Nomura, M.; Kumagai, N.; Iwamoto, S.; Ota, Y.; Arakawa, Y. Laser oscillation in a strongly coupled single-quantum-dot–nanocavity system. Nat. Phys. 2010, 6, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Volkov, Y. Quantum dots in nanomedicine: Recent trends, advances and unresolved issues. Biochem. Biophys. Res. Commun. 2015, 468, 419–427. [Google Scholar] [CrossRef] [Green Version]
- Gladkovskaya, O.; Gun’ko, Y.K.; O’connor, G.M.; Gogvadze, V.; Rochev, Y. In one harness: The interplay of cellular responses and subsequent cell fate after quantum dot uptake. Nanomedicine 2016, 11, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, W.F.; Cabrera, M.P.; Santos, N.R.M.; Napoleão, T.H.; Paiva, P.M.G.; Neves, R.P.; Silva, M.V.; Santos, B.S.; Coelho, L.C.B.B.; Cabral Filho, P.E.; et al. Evaluating glucose and mannose profiles in Candida species using quantum dots conjugated with Cramoll lectin as fluorescent nanoprobes. Microbiol. Res. 2020, 230, 126330. [Google Scholar] [CrossRef]
- Pagano, L.; Maestri, E.; Caldara, M.; White, J.C.; Marmiroli, N.; Marmiroli, M. Engineered Nanomaterial Activity at the Organelle Level: Impacts on the Chloroplasts and Mitochondria. ACS Sustain. Chem. Eng. 2018, 6, 12562–12579. [Google Scholar] [CrossRef]
- Rossi, R.; Ruotolo, R.; De Giorgio, G.; Marmiroli, M.; Villani, M.; Zappettini, A.; Marmiroli, N. Cadmium Sulfide Quantum Dots Adversely Affect Gametogenesis in Saccharomyces cerevisiae. Nanomaterials 2022, 12, 2208. [Google Scholar] [CrossRef] [PubMed]
- Levin, I.W.; Bhargava, R. Fourier transform infrared vibrational spectroscopic imaging: Integrating microscopy and molecular recognition. Annu. Rev. Phys. Chem. 2005, 56, 429–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petibois, C.; Déléris, G. Chemical mapping of tumor progression by FT-IR imaging: Towards molecular histopathology. Trends Biotechnol. 2006, 24, 455–462. [Google Scholar] [CrossRef]
- Siebert, F.; Hildebrandt, P. Vibrational Spectroscopy in Life Science; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2008. [Google Scholar]
- Griffith, P.R.; de Haseth, J.A. Fourier Transform Infrared Spectroscopy; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Berthomieu, C.; Hienerwadel, R. Fourier transform infrared (FTIR) spectroscopy. Photosynth. Res. 2009, 101, 157–170. [Google Scholar] [CrossRef]
- Ruotolo, R.; Marchini, G.; Ottonello, S. Membrane transporters and protein traffic networks differentially affecting metal tolerance: A genomic phenotyping study in yeast. Genome Biol. 2008, 9, R67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, S.; Ahting, U.; Eichacker, L.; Granvogl, B.; Go, N.E.; Nargang, F.; Neupert, W.; Nussberger, S. Role of Tom5 in Maintaining the Structural Stability of the TOM Complex of Mitochondria. J. Biol. Chem. 2005, 280, 14499–14506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habib, S.J.; Vasiljev, A.; Neupert, W.; Rapaport, D. Multiple functions of tail-anchor domains of mitochondrial outer membrane proteins. FEBS Lett. 2003, 555, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, V.; Srivastava, V.; Bulone, V.; Zappettini, A.; Villani, M.; Marmiroli, N.; Marmiroli, M. Proteomic Analysis Identifies Markers of Exposure to Cadmium Sulphide Quantum Dots (CdS QDs). Nanomaterials 2020, 10, 1214. [Google Scholar] [CrossRef]
- Pagano, L.; Marmiroli, M.; Villani, M.; Magnani, J.; Rossi, R.; Zappettini, A.; White, J.C.; Marmiroli, N. Engineered Nanomaterial Exposure Affects Organelle Genetic Material Replication in Arabidopsis thaliana. ACS Nano 2022, 16, 2249–2260. [Google Scholar] [CrossRef]
- Nedeva, T.; Petrova, V.; Hristozova, T.; Kujumdzieva, A. A Modified Procedure for Isolation of Yeast Mitochondrial DNA. Z. Naturforsh. 2002, 57, 960–962. [Google Scholar] [CrossRef]
- Birarda, G.; Bedolla, D.; Piccirilli, F.; Stani, C.; Vondracek, H.; Vaccari, L. Chemical analyses at micro and nano scale at SISSI-Bio beamline at Elettra-Sincrotrone Trieste. In Biomedical Vibrational Spectroscopy 2022: Advances in Research and Industry, Proceedings of the SPIE BiOS, San Francisco, CA, USA, 22–23 January 2022; SPIE: Bellingham, WA, USA, 2022; Volume 1195707. [Google Scholar]
- Barrientos, A.; Fontanesi, F.; Díaz, F. Evaluation of the Mitochondrial Respiratory Chain and Oxidative Phosphorylation System Using Polarography and Spectrophotometric Enzyme Assays. Curr. Protoc. Hum. Genet. 2009, 63, 19.3.1–19.3.14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta (BBA) Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Hellwig, P.; Scheide, D.; Bungert, S.; Mäntele, W.; Friedrich, T. FT-IR Spectroscopic Characterization of NADH:Ubiquinone Oxidoreductase (Complex I) from Escherichia coli: Oxidation of FeS Cluster N2 is Coupled with the Protonation of an Aspartate or Glutamate Side Chain. Biochemistry 2000, 39, 10884–10891. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, P.; Stolpe, S.; Friedrich, T. Fourier transform infrared spectroscopic study on the conformational reorganization inEscherichia coli complex I due to redox-driven proton translocation. Biopolymers 2004, 74, 69–72. [Google Scholar] [CrossRef]
- Hellwig, P. Infrared spectroscopic markers of quinones in proteins from the respiratory chain. Biochim. Biophys. Acta (BBA)-Bioenerg. 2015, 1847, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Baymann, F.; Robertson, D.E.; Dutton, P.L.; Mäntele, W. Electrochemical and spectroscopic investigations of the cytochrome bc1 complex from Rhodobacter capsulatus. Biochemistry 1999, 38, 13188–13199. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Anderka, O.; Ludwig, B.; Mäntele, W.; Hellwig, P. Electrochemical and FTIR Spectroscopic Characterization of the Cytochrome bc1 Complex from Paracoccus denitrificans: Evidence for Protonation Reactions Coupled to Quinone Binding. Biochemistry 2003, 42, 12391–12399. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.A.; Huang, L.-S.; Saechao, L.K.; Pon, N.G.; Valkova-Valchanova, M.; Daldal, F. X-Ray Structure of Rhodobacter Capsulatus Cytochrome bc1: Comparison with its Mitochondrial and Chloroplast Counterparts. Photosynth. Res. 2004, 81, 251–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaki, M.; Yakovlev, G.; Hirst, J.; Osyczka, A.; Dutton, P.L.; Marshall, D.; Rich, P.R. Direct observation of redox-linked histidine protonation changes in the iron-sulfur protein of the cytochrome bc1 complex by ATR-FTIR spectroscopy. Biochemistry 2005, 44, 4230–4237. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, M.; Osyczka, A.; Moser, C.C.; Dutton, P.L.; Rich, P.R.; April, R.V.; Re, V.; Recei, M.; May, V. ATR-FTIR Spectroscopy Studies of Iron—Sulfur Protein and Cytochrome c 1 in the Rhodobacter capsulatus Cytochrome bc 1 Complex. Biochemistry 2004, 43, 9477–9486. [Google Scholar] [CrossRef]
- Borer, P.; Hug, S.J. Photo-redox reactions of dicarboxylates and α-hydroxydicarboxylates at the surface of Fe(III)(hydr)oxides followed with in situ ATR-FTIR spectroscopy. J. Colloid Interface Sci. 2014, 416, 44–53. [Google Scholar] [CrossRef]
- Dietmeier, K.; Hönlinger, A.; Bömer, U.; Dekker, P.J.T.; Eckerskorn, C.; Lottspeich, F.; Kübrich, M.; Pfanner, N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 1997, 388, 195–200. [Google Scholar] [CrossRef]
- Pagano, L.; Caldara, M.; Villani, M.; Zappettini, A.; Marmiroli, N.; Marmiroli, M. In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae. Nanomaterials 2019, 9, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruotolo, R.; Pira, G.; Villani, M.; Zappettini, A.; Marmiroli, N. Ring-shaped corona proteins influence the toxicity of engineered nanoparticles to yeast. Environ. Sci. Nano 2018, 5, 1428–1440. [Google Scholar] [CrossRef]
- Naasz, S.; Altenburger, R.; Kühnel, D. Environmental mixtures of nanomaterials and chemicals: The Trojan-horse phenomenon and its relevance for ecotoxicity. Sci. Total. Environ. 2018, 635, 1170–1181. [Google Scholar] [CrossRef]
- Marmiroli, M.; Lepore, G.O.; Pagano, L.; D’Acapito, F.; Gianoncelli, A.; Villani, M.; Lazzarini, L.; White, J.C.; Marmiroli, N. The fate of CdS quantum dots in plants as revealed by extended X-ray absorption fine structure (EXAFS) analysis. Environ. Sci. Nano 2020, 7, 1150–1162. [Google Scholar] [CrossRef]
- Le, N.; Zhang, M.; Kim, K. Quantum Dots and Their Interaction with Biological Systems. Int. J. Mol. Sci. 2022, 23, 10763. [Google Scholar] [CrossRef] [PubMed]
- Paesano, L.; Vogli, M.; Marmiroli, M.; Bianchi, M.G.; Bussolati, O.; Zappettini, A.; Marmiroli, N. Cellular mechanisms of transcriptional regulation of human cell lines exposed to cadmium-based quantum dots. Environ. Sci. Nano 2023, 10, 1177–1189. [Google Scholar] [CrossRef]
- Mann, D.; Teuber, C.; Tennigkeit, S.A.; Schröter, G.; Gerwert, K.; Kötting, C. Mechanism of the intrinsic arginine finger in heterotrimeric G proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E8041–E8050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corte, L.; Tiecco, M.; Roscini, L.; Germani, R.; Cardinali, G. FTIR analysis of the metabolomic stress response induced by N-alkyltropinium bromide surfactants in the yeasts Saccharomyces cerevisiae and Candida albicans. Colloids Surf. B Biointerfaces 2014, 116, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Rossi, R.; White, J.C.; Marmiroli, N.; Marmiroli, M. Nanomaterials biotransformation: In planta mechanisms of action. Environ. Pollut. 2023, 318, 120834. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [Green Version]

| FTIR | Intact Cells | Isolated Mitochondria |
| Complex II | Major differences due to genetic background | Major differences due to effects of the treatment |
| Complex III | Major differences due to genetic background | - |
| Complex IV | Major differences due to genetic background | Major differences due to effects of the treatment |
| Enzymatic assay | Intact cells | Isolated mitochondria |
| Complex II | Major differences due to genetic background | Major differences due to effects of the treatment |
| Complex III | Major differences due to effects of the treatment | Major differences due to effects of the treatment |
| Complex IV | Major differences due to effects of the treatment | Major differences due to effects of the treatment |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marmiroli, M.; Birarda, G.; Gallo, V.; Villani, M.; Zappettini, A.; Vaccari, L.; Marmiroli, N.; Pagano, L. Cadmium Sulfide Quantum Dots, Mitochondrial Function and Environmental Stress: A Mechanistic Reconstruction through In Vivo Cellular Approaches in Saccharomyces cerevisiae. Nanomaterials 2023, 13, 1944. https://doi.org/10.3390/nano13131944
Marmiroli M, Birarda G, Gallo V, Villani M, Zappettini A, Vaccari L, Marmiroli N, Pagano L. Cadmium Sulfide Quantum Dots, Mitochondrial Function and Environmental Stress: A Mechanistic Reconstruction through In Vivo Cellular Approaches in Saccharomyces cerevisiae. Nanomaterials. 2023; 13(13):1944. https://doi.org/10.3390/nano13131944
Chicago/Turabian StyleMarmiroli, Marta, Giovanni Birarda, Valentina Gallo, Marco Villani, Andrea Zappettini, Lisa Vaccari, Nelson Marmiroli, and Luca Pagano. 2023. "Cadmium Sulfide Quantum Dots, Mitochondrial Function and Environmental Stress: A Mechanistic Reconstruction through In Vivo Cellular Approaches in Saccharomyces cerevisiae" Nanomaterials 13, no. 13: 1944. https://doi.org/10.3390/nano13131944