Cadmium Sulfide Quantum Dots Adversely Affect Gametogenesis in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of Nanoparticles
2.2. Yeast-Growth Conditions
2.3. Cytotoxicity and Sporulation Assays
2.4. Fluorescence Microscopy Analysis
2.5. Analysis of Spore Viability and Shape
2.6. Gene Expression Analysis
2.7. Flow Cytometry Analysis
2.8. In Vitro Protein-QD Binding Assay
2.9. Statistical Analysis
3. Results
3.1. Toxicity of CdS QDs in Different Growth Conditions
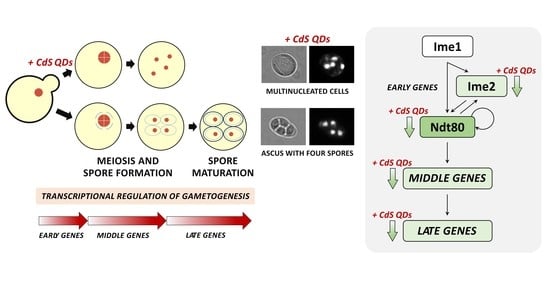
3.2. CdS QDs Strongly Affect the Sporulation Process
3.3. CdS QDs Adversely Affect Spore Germination and Morphology
3.4. CdS QDs Cause the Transcriptional Repression of Middle–Late Genes Involved in the Sporulation Process
3.5. Identification of Hard Corona Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardman, R. A toxicologic review of quantum dots: Toxicity depends on physicochemical and environmental factors. Environ. Health Perspect. 2006, 114, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Pietroiusti, A.; Stockmann-Juvala, H.; Lucaroni, F.; Savolainen, K. Nanomaterial exposure, toxicity, and impact on human health. WIREs Nanomed. Nanobiotechnol. 2018, 10, e1513. [Google Scholar] [CrossRef] [PubMed]
- Wegner, K.D.; Hildebrandt, N. Quantum dots: Bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem. Soc. Rev. 2015, 44, 4792–4834. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, M. Review of in vitro toxicological research of quantum dot and potentially involved mechanisms. Sci. Total Environ. 2018, 625, 940–962. [Google Scholar] [CrossRef]
- Cheki, M.; Moslehi, M.; Assadi, M. Marvelous applications of quantum dots. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 1141–1148. [Google Scholar]
- Ma-Hock, L.; Brill, S.; Wohlleben, W.; Farias, P.M.; Chaves, C.R.; Tenorio, D.P.; Fontes, A.; Santos, B.S.; Landsiedel, R.; Strauss, V.; et al. Short term inhalation toxicity of a liquid aerosol of CdS/Cd(OH)2 core shell quantum dots in male Wistar rats. Toxicol. Lett. 2012, 208, 115–124. [Google Scholar] [CrossRef]
- Peynshaert, K.; Soenen, S.J.; Manshian, B.B.; Doak, S.H.; Braeckmans, K.; De Smedt, S.C.; Remaut, K. Coating of Quantum Dots strongly defines their effect on lysosomal health and autophagy. Acta Biomater. 2017, 48, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, M. Dysfunction of various organelles provokes multiple cell death after quantum dot exposure. Int. J. Nanomed. 2018, 13, 2729–2742. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Tang, M. Review of the effects of manufactured nanoparticles on mammalian target organs. J. Appl. Toxicol. 2018, 38, 25–40. [Google Scholar] [CrossRef]
- Mei, J.; Yang, L.Y.; Lai, L.; Xu, Z.Q.; Wang, C.; Zhao, J.; Jin, J.C.; Jiang, F.L.; Liu, Y. The interactions between CdSe quantum dots and yeast Saccharomyces cerevisiae: Adhesion of quantum dots to the cell surface and the protection effect of ZnS shell. Chemosphere 2014, 112, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Maestri, E.; Caldara, M.; White, J.C.; Marmiroli, N.; Marmiroli, M. Engineered Nanomaterial Activity at the Organelle Level: Impacts on the Chloroplasts and Mitochondria. ACS Sustain. Chem. Eng. 2018, 6, 12562–12579. [Google Scholar] [CrossRef]
- Pasquali, F.; Agrimonti, C.; Pagano, L.; Zappettini, A.; Villani, M.; Marmiroli, M.; White, J.C.; Marmiroli, N. Nucleo-mitochondrial interaction of yeast in response to cadmium sulfide quantum dot exposure. J. Hazard. Mater. 2017, 324, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Shao, M.; Lai, L.; Liu, Y.; Xie, Z. Inhibition of autophagy contributes to the toxicity of cadmium telluride quantum dots in Saccharomyces cerevisiae. Int. J. Nanomed. 2016, 11, 3371–3383. [Google Scholar]
- Ruotolo, R.; Pira, G.; Villani, M.; Zappettini, A.; Marmiroli, N. Ring-shaped corona proteins influence the toxicity of engineered nanoparticles to yeast. Environ. Sci. Nano 2018, 5, 1428–1440. [Google Scholar] [CrossRef]
- Das, J.; Choi, Y.J.; Song, H.; Kim, J.H. Potential toxicity of engineered nanoparticles in mammalian germ cells and developing embryos: Treatment strategies and anticipated applications of nanoparticles in gene delivery. Hum. Reprod. Update 2016, 22, 588–619. [Google Scholar] [CrossRef] [Green Version]
- Talebi, A.R.; Khorsandi, L.; Moridian, M. The effect of zinc oxide nanoparticles on mouse spermatogenesis. J. Assist. Reprod. Genet. 2013, 30, 1203–1209. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Wang, L.; Yang, X.; Hu, Y.; Liu, Q. Acute Damage to the Sperm Quality and Spermatogenesis in Male Mice Exposed to Curcumin-Loaded Nanoparticles. Int. J. Nanomed. 2020, 15, 1853–1862. [Google Scholar] [CrossRef] [Green Version]
- Riaz Ahmed, K.B.; Nagy, A.M.; Brown, R.P.; Zhang, Q.; Malghan, S.G.; Goering, P.L. Silver nanoparticles: Significance of physicochemical properties and assay interference on the interpretation of in vitro cytotoxicity studies. Toxicol. Vitr. 2017, 38, 179–192. [Google Scholar] [CrossRef]
- Blum, J.L.; Edwards, J.R.; Prozialeck, W.C.; Xiong, J.Q.; Zelikoff, J.T. Effects of Maternal Exposure to Cadmium Oxide Nanoparticles During Pregnancy on Maternal and Offspring Kidney Injury Markers Using a Murine Model. J. Toxicol. Environ. Health A 2015, 78, 711–724. [Google Scholar] [CrossRef] [Green Version]
- Sekigawa, M.; Kunoh, T.; Wada, S.; Mukai, Y.; Ohshima, K.; Ohta, S.; Goshima, N.; Sasaki, R.; Mizukami, T. Comprehensive screening of human genes with inhibitory effects on yeast growth and validation of a yeast cell-based system for screening chemicals. J. Biomol. Screen. 2010, 15, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, J.M.; Young, J.H.; Kachroo, A.H.; Marcotte, E.M. Efforts to make and apply humanized yeast. Brief. Funct. Genom. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sopko, R.; Raithatha, S.; Stuart, D. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 2002, 22, 7024–7040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, D.; Yang, Y.; Lacefield, S. Positive feedback of NDT80 expression ensures irreversible meiotic commitment in budding yeast. PLoS Genet. 2014, 10, e1004398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pak, J.; Segall, J. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2002, 22, 6417–6429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, H.; Namdar, M.; Ganier, O.; Gregan, J.; Mechali, M.; Kearsey, S.E. Sequential steps in DNA replication are inhibited to ensure reduction of ploidy in meiosis. Mol. Biol. Cell 2013, 24, 578–587. [Google Scholar] [CrossRef]
- Phizicky, D.V.; Berchowitz, L.E.; Bell, S.P. Multiple kinases inhibit origin licensing and helicase activation to ensure reductive cell division during meiosis. eLife 2018, 7, e33309. [Google Scholar] [CrossRef]
- Villani, M.; Calestani, D.; Lazzarini, L.; Zanotti, L.; Mosca, R.; Zappettini, A. Extended functionality of ZnO nanotetrapods by solution-based coupling with CdS nanoparticles. J. Mater. Chem. 2012, 22, 5694–5699. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.; Zhu, H.; Hadjipanayis, G.C.; Xiao, J.Q. Low-temperature synthesis of hexagonal (Wurtzite) ZnS nanocrystals. J. Am. Chem. Soc. 2004, 126, 6874–6875. [Google Scholar] [CrossRef]
- Parodi, E.M.; Baker, C.S.; Tetzlaff, C.; Villahermosa, S.; Huang, L.S. SPO71 mediates prospore membrane size and maturation in Saccharomyces cerevisiae. Eukaryot. Cell 2012, 11, 1191–1200. [Google Scholar] [CrossRef] [Green Version]
- Parodi, E.M.; Roesner, J.M.; Huang, L.S. SPO73 and SPO71 Function Cooperatively in Prospore Membrane Elongation during Sporulation in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0143571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, C.M.; Wagner, M.; Dunham, M.J.; Shin, M.E.; Ahmed, N.T.; Winter, E. The Ras/cAMP pathway and the CDK-like kinase Ime2 regulate the MAPK Smk1 and spore morphogenesis in Saccharomyces cerevisiae. Genetics 2009, 181, 511–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, M.; Briza, P.; Pierce, M.; Winter, E. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 1999, 151, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, T.; Sarkar, D.; Mashayekhi, H.; Xing, B. Growth and enzymatic activity of maize (Zea mays L.) plant: Solution culture test for copper dioxide nano particles. J. Plant Nutr. 2016, 39, 99–115. [Google Scholar] [CrossRef]
- Grigore, M.E.; Biscu, E.R.; Holban, A.M.; Gestal, M.C.; Grumezescu, A.M. Methods of Synthesis, Properties and Biomedical Applications of CuO Nanoparticles. Pharmaceuticals 2016, 9, 75. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, Y.; Xu, Y.; Yin, Y.; Guo, H.; Du, W. Divergence in response of lettuce (var. ramosa Hort.) to copper oxide nanoparticles/microparticles as potential agricultural fertilizer. Environ. Pollut. Bioavailab. 2019, 31, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, H.L.; Gustafsson, J.; Cronholm, P.; Moller, L. Size-dependent toxicity of metal oxide particles—A comparison between nano- and micrometer size. Toxicol. Lett. 2009, 188, 112–118. [Google Scholar] [CrossRef]
- Kasemets, K.; Suppi, S.; Kunnis-Beres, K.; Kahru, A. Toxicity of CuO nanoparticles to yeast Saccharomyces cerevisiae BY4741 wild-type and its nine isogenic single-gene deletion mutants. Chem. Res. Toxicol. 2013, 26, 356–367. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pagano, L.; Rossi, R.; De La Torre-Roche, R.; Lepore, G.O.; Ruotolo, R.; Gariani, G.; Bonanni, V.; Pollastri, S.; Puri, A.; et al. Copper Oxide Nanomaterial Fate in Plant Tissue: Nanoscale Impacts on Reproductive Tissues. Environ. Sci. Technol. 2021, 55, 10769–10783. [Google Scholar] [CrossRef]
- Ruotolo, R.; De Giorgio, G.; Minato, I.; Bianchi, M.G.; Bussolati, O.; Marmiroli, N. Cerium Oxide Nanoparticles Rescue alpha-Synuclein-Induced Toxicity in a Yeast Model of Parkinson’s Disease. Nanomaterials 2020, 10, 235. [Google Scholar] [CrossRef] [Green Version]
- Marmiroli, M.; Pagano, L.; Savo Sardaro, M.L.; Villani, M.; Marmiroli, N. Genome-wide approach in Arabidopsis thaliana to assess the toxicity of cadmium sulfide quantum dots. Environ. Sci. Technol. 2014, 48, 5902–5909. [Google Scholar] [CrossRef] [PubMed]
- Pagano, L.; Marmiroli, M.; Villani, M.; Magnani, J.; Rossi, R.; Zappettini, A.; White, J.C.; Marmiroli, N. Engineered Nanomaterial Exposure Affects Organelle Genetic Material Replication in Arabidopsis thaliana. ACS Nano 2022, 16, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmiroli, N.; Ferri, M.; Puglisi, P.P. Involvement of mitochondrial protein synthesis in sporulation: Effects of erythromycin on macromolecular synthesis, meiosis, and ascospore formation in Saccharomyces cerevisiae. J. Bacteriol. 1983, 154, 118–129. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, Q.; Liu, C.; Shang, Y.; Wen, F.; Wang, F.; Liu, W.; Xiao, W.; Li, W. A Role for the Respiratory Chain in Regulating Meiosis Initiation in Saccharomyces cerevisiae. Genetics 2018, 208, 1181–1194. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Sakakibara, A.; Yamasaki, M.; Yoda, K. Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J. Biol. Chem. 1997, 272, 16308–16314. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.; Bannigan, J. Cadmium: Toxic effects on the reproductive system and the embryo. Reprod. Toxicol. 2008, 25, 304–315. [Google Scholar] [CrossRef]
- Geng, H.X.; Wang, L. Cadmium: Toxic effects on placental and embryonic development. Environ. Toxicol. Pharmacol. 2019, 67, 102–107. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef]
- Chu, S.; Herskowitz, I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1998, 1, 685–696. [Google Scholar] [CrossRef]
- Chu, S.; DeRisi, J.; Eisen, M.; Mulholland, J.; Botstein, D.; Brown, P.O.; Herskowitz, I. The transcriptional program of sporulation in budding yeast. Science 1998, 282, 699–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, E.M.; Joshi, P.R.; Jorgensen, V.; Yunus, J.; Berchowitz, L.E.; Unal, E. Developmental regulation of an organelle tether coordinates mitochondrial remodeling in meiosis. J. Cell Biol. 2019, 218, 559–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamoureux, J.S.; Stuart, D.; Tsang, R.; Wu, C.; Glover, J.N. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 2002, 21, 5721–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Gene Name | Primer Sequence (5′–3′) | Amplicon Size (bp) |
|---|---|---|
| ACT1 | FW: GAGGTTGCTGCTTTGGTTATTGA RE: CGTCGTCACCGGCAAAA | 67 |
| IME1 | FW: CGTTGAAAAATCACCACCGCCA RE: CTGAAGGAGTAAGCCGCAGCA | 111 |
| IME2 | FW: ACGGCCTACGTTTCCACAAGAT RE: CCACGCACCCGAATGCCCAA | 100 |
| NDT80 | FW: GCCATCAATGGCGCAGCCGT RE: CGAGATGGAGGCCCCAGAGT | 106 |
| SMK1 | FW: TGACCAGCTCGCCCTATGACG RE: CCGAGAGCTGCACGGACGAAT | 93 |
| SPO1 | FW: TGGATTATCAGGCGGAAGTTGG RE: TCCTCTTCAAGGTCCCACTCTT | 93 |
| RIM4 | FW: GGCAAAACATTTACAGGGCCAG RE: GCTTTCCTGCTGGGATCCGC | 90 |
| DIT1 | FW: GGTCGATGATGACGTCGTGAG RE: AGCCAATGGCGTCAACACCAG | 89 |
| DIT2 | FW: CGTGCAAGTTGGGGGCGGAA RE: GCCCCAAGTTTTGGGATCGTG | 92 |
| SPO20 | FW: TCACCCAAACTGTCGGTTCGATGA RE: GTAGCAAGGCCATCCCTTTCG | 90 |
| SPS100 | FW: ACGCGGAAGGTAGAGGCACTT RE: CCTGTGGGCGTTTTGTCTGGT | 100 |
| Protein | Description (UniProt Accession N.) | Score 1 | Coverage 2 | Molecular Weight (KDa) | pI | Biological Process |
|---|---|---|---|---|---|---|
| Atp1 | Alpha subunit of the mitochondrial ATP synthase (P07251) | 132.45 | 52% | 59 | 9.5 | Energy metabolism |
| Cdc19 | Pyruvate kinase (P00549) | 192.37 | 83% | 55 | 7.8 | Energy metabolism |
| Idp2 | Isocitrate dehydrogenase (P41939) | 174.19 | 77% | 47 | 6.1 | Energy metabolism |
| Lat1 | Dihydrolipoamide acetyltransferase component of the pyruvate dehydrogenase complex (P12695) | 79.57 | 38% | 52 | 8.0 | Energy metabolism |
| Pdb1 | E1 beta subunit of the pyruvate dehydrogenase complex (P32473) | 73.89 | 36% | 40 | 5.0 | Energy metabolism |
| Tdh2 | Glyceraldehyde-3-phosphate dehydrogenase (P00358) | 137.4 | 55% | 36 | 7.0 | Energy metabolism |
| Tdh3 | Glyceraldehyde-3-phosphate dehydrogenase (P00359) | 149.31 | 72% | 36 | 7.0 | Energy metabolism |
| Leu2 | Beta-isopropylmalate dehydrogenase (P04173) | 81.46 | 58% | 39 | 5.5 | Metabolic process |
| Psa1 | Mannose-1-phosphate guanyltransferase (P41940) | 73.16 | 37% | 40 | 6.3 | Metabolic process |
| EF-1α (Tef1/2) | Translational elongation factor EF-1 alpha (P02994) | 93.01 | 44% | 50 | 9.1 | Translation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, R.; Ruotolo, R.; De Giorgio, G.; Marmiroli, M.; Villani, M.; Zappettini, A.; Marmiroli, N. Cadmium Sulfide Quantum Dots Adversely Affect Gametogenesis in Saccharomyces cerevisiae. Nanomaterials 2022, 12, 2208. https://doi.org/10.3390/nano12132208
Rossi R, Ruotolo R, De Giorgio G, Marmiroli M, Villani M, Zappettini A, Marmiroli N. Cadmium Sulfide Quantum Dots Adversely Affect Gametogenesis in Saccharomyces cerevisiae. Nanomaterials. 2022; 12(13):2208. https://doi.org/10.3390/nano12132208
Chicago/Turabian StyleRossi, Riccardo, Roberta Ruotolo, Giuseppe De Giorgio, Marta Marmiroli, Marco Villani, Andrea Zappettini, and Nelson Marmiroli. 2022. "Cadmium Sulfide Quantum Dots Adversely Affect Gametogenesis in Saccharomyces cerevisiae" Nanomaterials 12, no. 13: 2208. https://doi.org/10.3390/nano12132208