Detection of C-Reactive Protein Using Histag-HRP Functionalized Nanoconjugate with Signal Amplified Immunoassay
Abstract
:1. Introduction
2. Experimental Procedure
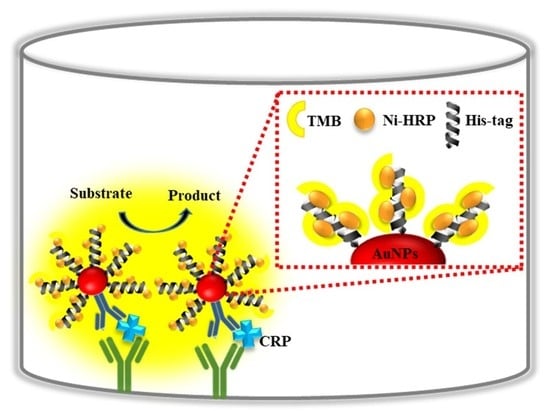
2.1. Preparation of the 6XHis-Gold-HRP Nanoprobes
2.2. Estimation of 6XHis-HRP Conjugated on a Gold Nanoparticle
2.3. Enzyme-6Xhis Nanoprobe Immunoassay for the Detection of CRP
2.4. Conventional Enzyme-Linked Immunosorbent Assay (ELISA) for CRP
3. Result and Discussions
3.1. Development of HRP-Labeled 6Xhis Nanoprobe
3.2. Development of Immunoassay
3.3. Analytical Performance of Nanoprobe-ELISA for CRP Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dillenseger, L.; Langlet, C.; Iacobelli, S.; Lavaux, T.; Ratomponirina, C.; Labenne, M.; Astruc, D.; Severac, F.; Gouyon, J.B.; Kuhn, P. Early Inflammatory Markers for the Diagnosis of Late-Onset Sepsis in Neonates: The Nosodiag Study. Front. Pediatr. 2018, 6, 346. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tan, G.-J.; Han, L.-N.; Bai, Y.-Y.; He, M.; Liu, H.-B. Novel biomarkers for cardiovascular risk prediction. J. Geriatr. Cardiol. 2017, 14, 135–150. [Google Scholar]
- May, A.; Wang, T.J. Evaluating the role of biomarkers for cardiovascular risk prediction: Focus on CRP, BNP and urinary microalbumin. Expert Rev. Mol. Diagn. 2007, 7, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Vance, S.A.; Sandros, M.G. Zeptomole Detection of C-Reactive Protein in Serum by a Nanoparticle Amplified Surface Plasmon Resonance Imaging Aptasensor. Sci. Rep. 2014, 4, 5129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vashist, S.K.; Venkatesh, A.G.; Marion Schneider, E.; Beaudoin, C.; Luppa, P.B.; Luong, J.H.T. Bioanalytical advances in assays for C-reactive protein. Biotechnol. Adv. 2016, 34, 272–290. [Google Scholar] [CrossRef] [PubMed]
- Dorraki, M.; Fouladzadeh, A.; Salamon, S.J.; Allison, A.; Coventry, B.J.; Abbott, D. On detection of periodicity in C-reactive protein (CRP) levels. Sci. Rep. 2018, 8, 11979. [Google Scholar] [CrossRef] [PubMed]
- Otsuji, S.; Shibata, H.; Umeda, M. Turbidimetric immunoassay of serum C-reactive protein. Clin. Chem. 1982, 28, 2121–2124. [Google Scholar] [CrossRef] [PubMed]
- Sarikaputi, M.; Morimatsu, M.; Yamamoto, S.; Syuto, B.; Saito, M.; Naiki, M. Latex agglutination test: A simple, rapid and practical method for bovine serum CRP determination. Jpn. J. Vet. Res. 1992, 40, 1–12. [Google Scholar]
- Barka, N.; Tomasi, J.P.; Stadtsbaeder, S. Use of whole Streptococcus pneumoniae cells as a solid phase sorbent for C-reactive protein measurement by ELISA. J. Immunol. Methods 1985, 82, 57–63. [Google Scholar] [CrossRef]
- Wu, T.L.; Tsai, I.C.; Chang, P.Y.; Tsao, K.C.; Sun, C.F.; Wu, L.L.; Wu, J.T. Establishment of an in-house ELISA and the reference range for serum amyloid A (SAA). Complementarity between SAA and C-reactive protein as markers of inflammation. Clin. Chim. Acta 2007, 376, 72–76. [Google Scholar] [CrossRef]
- Zhao, L.J.; Yu, R.J.; Ma, W.; Han, H.X.; Tian, H.; Qian, R.C.; Long, Y.T. Sensitive detection of protein biomarkers using silver nanoparticles enhanced immunofluorescence assay. Theranostics 2017, 7, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Alhabbab, R.Y. Precipitation and Agglutination Reactions. In Basic Serological Testing; Springer: Berlin, Germany, 2018; pp. 23–30. [Google Scholar]
- De Ávila BE, F.; Escamilla-Gómez, V.; Campuzano, S.; Pedrero, M.; Salvador, J.P.; Marco, M.P.; Pingarrón, J.M. Ultrasensitive amperometric magnetoimmunosensor for human C-reactive protein quantification in serum. Sens. Actuators B Chem. 2013, 188, 212–220. [Google Scholar]
- Baldini, F.; Giannetti, A.; Senesi, F.; Trono, C.; Bolzoni, L.; Porro, G. An Optical Platform Based on Fluorescence Anisotropy for C Reactive Protein and Procalcitonine Assay. In Sensors and Microsystems; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Vashist, S.K.; Marion Schneider, E.; Lam, E.; Hrapovic, S.; Luong, J.H.T. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci. Rep. 2014, 4, 4407. [Google Scholar] [CrossRef] [PubMed]
- Lakshmipriya, T.; Gopinath, S.C.B.; Tang, T.-H. Biotin-Streptavidin Competition Mediates Sensitive Detection of Biomolecules in Enzyme Linked Immunosorbent Assay. PLoS ONE 2016, 11, e0151153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Garcia-D’Angeli, A.; Brennan, J.P.; Huo, Q. Predicting detection limits of enzyme-linked immunosorbent assay (ELISA) and bioanalytical techniques in general. Analyst 2014, 139, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Vdovenko, M.M.; Byzova, N.A.; Zherdev, A.V.; Dzantiev, B.B.; Sakharov, I.Y. Ternary covalent conjugate (antibody–gold nanoparticle–peroxidase) for signal enhancement in enzyme immunoassay. RSC Adv. 2016, 6, 48827–48833. [Google Scholar] [CrossRef]
- O’Sullivan, M.J.; Marks, V. Methods for the preparation of enzyme-antibody conjugates for use in enzyme immunoassay. Methods Enzymol. 1981, 73, 147–166. [Google Scholar]
- Green, N.M. Avidin and streptavidin. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1990; Volume 184, pp. 51–67. [Google Scholar]
- Zhou, W.; Gao, X.; Liu, D.; Chen, X. Gold Nanoparticles for In Vitro Diagnostics. Chem. Rev. 2015, 115, 10575–10636. [Google Scholar] [CrossRef] [Green Version]
- Hayat, M.A. Colloidal Gold: Principles, Methods, and Applications; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Han, K.-C.; Yang, E.G.; Ahn, D.-R. A highly sensitive, multiplex immunoassay using gold nanoparticle-enhanced signal amplification. Chem. Commun. 2012, 48, 5895–5897. [Google Scholar] [CrossRef]
- Zhang, P.; Cui, Y.; Anderson, C.F.; Zhang, C.; Li, Y.; Wang, R.; Cui, H. Peptide-based nanoprobes for molecular imaging and disease diagnostics. Chem. Soc. Rev. 2018, 47, 3490–3529. [Google Scholar] [CrossRef]
- Xiao, X.; Kuang, Z.; Slocik, J.M.; Tadepalli, S.; Brothers, M.; Kim, S.; Mirau, P.A.; Butkus, C.; Farmer, B.L.; Hall, C.K.; et al. Advancing Peptide-Based Biorecognition Elements for Biosensors Using in-Silico Evolution. ACS Sens. 2018, 3, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Ghafoor, F.; Iqbal, I.A.; Saqlan Naqvi, S.M. Development of a high sensitivity C-reactive protein immunoassay and comparison with a commercial kit. J. Immunoass. Immunochem. 2017, 38, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Unten, S.K.; Hokama, Y. Enzyme immunoassay for c-reactive protein analysis. J. Clin. Lab. Anal. 1987, 1, 136–139. [Google Scholar] [CrossRef]
- Salonen, E.-M. A rapid and sensitive solid-phase enzyme immunoassay for C-reactive protein. J. Immunol. Methods 1982, 48, 45–50. [Google Scholar] [CrossRef]
- Stein, W.H.; Moore, S. Amino acid composition of β-lactoglobulin and bovine serum albumin. J. Biol. Chem. 1949, 178, 79–91. [Google Scholar]
- Rowinska-Zyrek, M.; Witkowska, D.; Potocki, S.; Remelli, M.; Kozlowski, H. His-rich sequences–is plagiarism from nature a good idea? New J. Chem. 2013, 37, 58–70. [Google Scholar] [CrossRef]
- Valenti, L.E.; De Pauli, C.P.; Giacomelli, C.E. The binding of Ni(II) ions to hexahistidine as a model system of the interaction between nickel and His-tagged proteins. J. Inorg. Biochem. 2006, 100, 192–200. [Google Scholar] [CrossRef]
- Ambrosi, A.; Castaneda, M.T.; Killard, A.J.; Smyth, M.R.; Alegret, S.; Merkoci, A. Double-codified gold nanolabels for enhanced immunoanalysis. Anal. Chem. 2007, 79, 5232–5240. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV−Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Thobhani, S.; Attree, S.; Boyd, R.; Kumarswami, N.; Noble, J.; Szymanski, M.; Porter, R.A. Bioconjugation and characterisation of gold colloid-labelled proteins. J. Immunol. Methods 2010, 356, 60–69. [Google Scholar] [CrossRef]
- Pollitt, M.J.; Buckton, G.; Piper, R.; Brocchini, S. Measuring antibody coatings on gold nanoparticles by optical spectroscopy. RSC Adv. 2015, 5, 24521–24527. [Google Scholar] [CrossRef]
- Madaboosi, N.; Soares, R.R.G.; Chu, V.; Conde, J.P. A microfluidic immunoassay platform for the detection of free prostate specific antigen: A systematic and quantitative approach. Analyst 2015, 140, 4423–4433. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.L.; Devanarayan, V.; Kriauciunas, A.; Manetta, J.; Montrose, C.; Sittampalam, S. Immunoassay methods. In Assay Guidance Manual [Internet]; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2014. [Google Scholar]
- Waritani, T.; Chang, J.; McKinney, B.; Terato, K. An ELISA protocol to improve the accuracy and reliability of serological antibody assays. MethodsX 2017, 4, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dzakah, E.E.; Kang, Y.; Cai, Y.; Wu, P.; Tang, B.; Li, R.; He, X. A sensitive and rapid chemiluminescence immunoassay for point-of-care testing (POCT) of copeptin in serum based on high-affinity monoclonal antibodies via cytokine-assisted immunization. Int. J. Nanomed. 2019, 14, 4293–4307. [Google Scholar] [CrossRef] [Green Version]
- Retout, M.; Valkenier, H.; Triffaux, E.; Doneux, T.; Bartik, K.; Bruylants, G. Rapid and Selective Detection of Proteins by Dual Trapping Using Gold Nanoparticles Functionalized with Peptide Aptamers. ACS Sens. 2016, 1, 929–933. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Chen, C.-P.; Wu, T.-H.; Yang, C.-H.; Lin, C.-W.; Chen, C.-Y. Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials 2019, 9, 861. [Google Scholar] [CrossRef] [Green Version]







| Factors | Developed | Conventional |
|---|---|---|
| R2 | 0.984 | 0.982 |
| Slope | 0.669 | 0.184 |
| Intercept | 0.0039 | −0.001 |
| LOD (ng/mL) | 0.032 | 0.402 |
| Assay Time | <2 h | >4 h |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqui, M.F.; Khan, Z.A.; Park, S. Detection of C-Reactive Protein Using Histag-HRP Functionalized Nanoconjugate with Signal Amplified Immunoassay. Nanomaterials 2020, 10, 1240. https://doi.org/10.3390/nano10061240
Siddiqui MF, Khan ZA, Park S. Detection of C-Reactive Protein Using Histag-HRP Functionalized Nanoconjugate with Signal Amplified Immunoassay. Nanomaterials. 2020; 10(6):1240. https://doi.org/10.3390/nano10061240
Chicago/Turabian StyleSiddiqui, Mohd Farhan, Zeeshan Ahmad Khan, and Seungkyung Park. 2020. "Detection of C-Reactive Protein Using Histag-HRP Functionalized Nanoconjugate with Signal Amplified Immunoassay" Nanomaterials 10, no. 6: 1240. https://doi.org/10.3390/nano10061240