Plasma microRNA Profiling Reveals Novel Biomarkers of Epicardial Adipose Tissue: A Multidetector Computed Tomography Study
Abstract
:1. Introduction
2. Experimental Section
2.1. Study Population
2.2. Coronary Computed Tomography Angiography (CCTA)
2.3. Blood Collection
2.4. Epicardial Adipose Tissue (EAT)
2.5. High-Sensitive C-Reactive Protein (CRP) Concentration
2.6. MicroRNA Isolation
2.7. Quantification of MicroRNA
2.8. Statistical Analysis
3. Results
3.1. Study Population
3.2. Profiling of Plasma MicroRNAs
3.3. Plasma MicroRNAs and Epicardial Fat Volume
3.4. Performance of Plasma MicroRNAs as Biomarkers of Epicardial Fat Volume
3.5. Validation Using Alternative Cutoffs of Epicardial Fat Volume
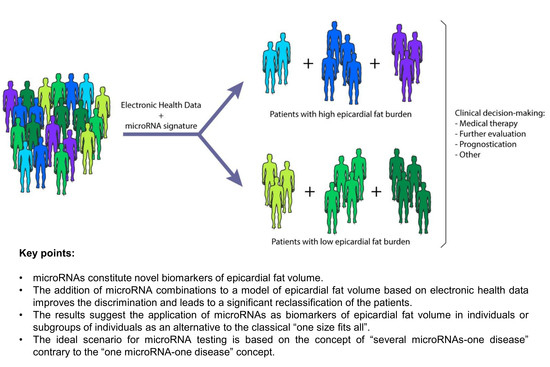
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- De Gonzalo-Calvo, D.; Vea, A.; Bar, C.; Fiedler, J.; Couch, L.S.; Brotons, C.; Llorente-Cortes, V.; Thum, T. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: A novel tool for personalized medicine? Eur. Heart J. 2019, 40, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Nicassio, F.; Marzi, M.; Belloni, E.; Dall’olio, V.; Bernard, L.; Pelosi, G.; Maisonneuve, P.; Veronesi, G.; Di Fiore, P.P. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol. Med. 2011, 3, 495–503. [Google Scholar] [CrossRef]
- Ralfkiaer, U.; Hagedorn, P.H.; Bangsgaard, N.; Lovendorf, M.B.; Ahler, C.B.; Svensson, L.; Kopp, K.L.; Vennegaard, M.T.; Lauenborg, B.; Zibert, J.R.; et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011, 118, 5891–5900. [Google Scholar] [CrossRef]
- De Gonzalo-Calvo, D.; Iglesias-Gutierrez, E.; Llorente-Cortes, V. Epigenetic Biomarkers and Cardiovascular Disease: Circulating MicroRNAs. Rev. Esp. Cardiol. 2017, 70, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Regazzi, R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacobellis, G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat. Rev. Endocrinol. 2015, 11, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Packer, M. The epicardial adipose inflammatory triad: Coronary atherosclerosis, atrial fibrillation, and heart failure with a preserved ejection fraction. Eur. J. Heart Fail. 2018, 20, 1567–1569. [Google Scholar] [CrossRef]
- Ansaldo, A.M.; Montecucco, F.; Sahebkar, A.; Dallegri, F.; Carbone, F. Epicardial adipose tissue and cardiovascular diseases. Int. J. Cardiol. 2019, 278, 254–260. [Google Scholar] [CrossRef]
- Blumensatt, M.; Fahlbusch, P.; Hilgers, R.; Bekaert, M.; Herzfeld de Wiza, D.; Akhyari, P.; Ruige, J.B.; Ouwens, D.M. Secretory products from epicardial adipose tissue from patients with type 2 diabetes impair mitochondrial beta-oxidation in cardiomyocytes via activation of the cardiac renin-angiotensin system and induction of miR-208a. Basic Res. Cardiol. 2017, 112, 2. [Google Scholar] [CrossRef]
- Mancio, J.; Azevedo, D.; Saraiva, F.; Azevedo, A.I.; Pires-Morais, G.; Leite-Moreira, A.; Falcao-Pires, I.; Lunet, N.; Bettencourt, N. Epicardial adipose tissue volume assessed by computed tomography and coronary artery disease: A systematic review and meta-analysis. Eur. Heart J. Cardiovasc. Imaging 2018, 19, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Nyman, K.; Graner, M.; Pentikainen, M.O.; Lundbom, J.; Hakkarainen, A.; Siren, R.; Nieminen, M.S.; Taskinen, M.R.; Lundbom, N.; Lauerma, K. Cardiac steatosis and left ventricular function in men with metabolic syndrome. J. Cardiovasc. Magn. Reson. 2013, 15, 103. [Google Scholar] [CrossRef]
- Bos, D.; Vernooij, M.W.; Shahzad, R.; Kavousi, M.; Hofman, A.; van Walsum, T.; Deckers, J.W.; Ikram, M.A.; Heeringa, J.; Franco, O.H.; et al. Epicardial Fat Volume and the Risk of Atrial Fibrillation in the General Population Free of Cardiovascular Disease. JACC Cardiovasc. Imaging 2017, 10, 1405–1407. [Google Scholar] [CrossRef]
- Spearman, J.V.; Renker, M.; Schoepf, U.J.; Krazinski, A.W.; Herbert, T.L.; De Cecco, C.N.; Nietert, P.J.; Meinel, F.G. Prognostic value of epicardial fat volume measurements by computed tomography: A systematic review of the literature. Eur. Radiol. 2015, 25, 3372–3381. [Google Scholar] [CrossRef] [PubMed]
- Pierdomenico, S.D.; Pierdomenico, A.M.; Cuccurullo, F.; Iacobellis, G. Meta-analysis of the relation of echocardiographic epicardial adipose tissue thickness and the metabolic syndrome. Am. J. Cardiol. 2013, 111, 73–78. [Google Scholar] [CrossRef]
- Beltowski, J. Epicardial adipose tissue: The new target for statin therapy. Int. J. Cardiol. 2019, 274, 353–354. [Google Scholar] [CrossRef]
- Iacobellis, G.; Mohseni, M.; Bianco, S.D.; Banga, P.K. Liraglutide causes large and rapid epicardial fat reduction. Obesity 2017, 25, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gonzalo-Calvo, D.; Vilades, D.; Nasarre, L.; Carreras, F.; Leta, R.; Garcia-Moll, X.; Llorente-Cortes, V. Circulating levels of soluble low-density lipoprotein receptor-related protein 1 (sLRP1) as novel biomarker of epicardial adipose tissue. Int. J. Cardiol. 2016, 223, 371–373. [Google Scholar] [CrossRef]
- Gonzalo-Calvo, D.; Colom, C.; Vilades, D.; Rivas-Urbina, A.; Moustafa, A.H.; Perez-Cuellar, M.; Sanchez-Quesada, J.L.; Perez, A.; Llorente-Cortes, V. Soluble LRP1 is an independent biomarker of epicardial fat volume in patients with type 1 diabetes mellitus. Sci. Rep. 2018, 8, 1054. [Google Scholar] [CrossRef] [Green Version]
- Shmilovich, H.; Dey, D.; Cheng, V.Y.; Rajani, R.; Nakazato, R.; Otaki, Y.; Nakanishi, R.; Slomka, P.J.; Thomson, L.E.; Hayes, S.W.; et al. Threshold for the upper normal limit of indexed epicardial fat volume: Derivation in a healthy population and validation in an outcome-based study. Am. J. Cardiol. 2011, 108, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
- Tuck, M.K.; Chan, D.W.; Chia, D.; Godwin, A.K.; Grizzle, W.E.; Krueger, K.E.; Rom, W.; Sanda, M.; Sorbara, L.; Stass, S.; et al. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J. Proteome Res. 2009, 8, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 2014, 11, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B., Sr.; Steyerberg, E.W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat. Med. 2011, 30, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Harrell, F.E. Regression Modeling Strategies. Springer Series in Statistics; Springer: Cham, Germany; New York, NY, USA, 2015. [Google Scholar]
- Kass, G. An exploratory technique for investigating large quantities of categorical data. Appl. Stat. 1980, 29, 119–127. [Google Scholar] [CrossRef]
- Chen, Y.; Buyel, J.J.; Hanssen, M.J.; Siegel, F.; Pan, R.; Naumann, J.; Schell, M.; van der Lans, A.; Schlein, C.; Froehlich, H.; et al. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat. Commun. 2016, 7, 11420. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; You, L.; Zhu, L.; Wang, X.; Zhou, Y.; Li, Y.; Wen, J.; Xia, Y.; Wang, X.; Ji, C.; et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism 2018, 78, 95–105. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; McAnena, O.J.; O’Brien, T.; Kerin, M.J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J. Clin. Endocrinol. Metab. 2011, 96, E846–E850. [Google Scholar] [CrossRef]
- Pek, S.L.; Sum, C.F.; Lin, M.X.; Cheng, A.K.; Wong, M.T.; Lim, S.C.; Tavintharan, S. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Mol. Cell Endocrinol. 2016, 427, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; Lopez-Bermejo, A.; Fernandez-Real, J.M. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; van der Meer, R.W.; Rijzewijk, L.J.; Smit, J.W.; Revuelta-Lopez, E.; Nasarre, L.; Escola-Gil, J.C.; Lamb, H.J.; Llorente-Cortes, V. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci. Rep. 2017, 7, 47. [Google Scholar] [CrossRef]
- Nakazato, R.; Rajani, R.; Cheng, V.Y.; Shmilovich, H.; Nakanishi, R.; Otaki, Y.; Gransar, H.; Slomka, P.J.; Hayes, S.W.; Thomson, L.E.; et al. Weight change modulates epicardial fat burden: A 4-year serial study with non-contrast computed tomography. Atherosclerosis 2012, 220, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Parisi, V.; Petraglia, L.; D’Esposito, V.; Cabaro, S.; Rengo, G.; Caruso, A.; Grimaldi, M.G.; Baldascino, F.; De Bellis, A.; Vitale, D.; et al. Statin therapy modulates thickness and inflammatory profile of human epicardial adipose tissue. Int. J. Cardiol. 2019, 274, 326–330. [Google Scholar] [CrossRef] [Green Version]
- Walter, E.; Dellago, H.; Grillari, J.; Dimai, H.P.; Hackl, M. Cost-utility analysis of fracture risk assessment using microRNAs compared with standard tools and no monitoring in the Austrian female population. Bone 2018, 108, 44–54. [Google Scholar] [CrossRef]
- Packer, M. Epicardial Adipose Tissue May Mediate Deleterious Effects of Obesity and Inflammation on the Myocardium. J. Am. Coll. Cardiol. 2018, 71, 2360–2372. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014, 124, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Qin, S.; Li, W.; Wu, W.; Yang, J.; Chu, M.; Li, X.; Huo, Y.; Schaer, G.L.; Wang, S.; et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J. Am. Coll. Cardiol. 2015, 65, 2526–2537. [Google Scholar] [CrossRef]
- Bär, C.; Thum, T.; de Gonzalo-Calvo, D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics 2019, 11, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARgamma. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Alimujiang, M.; Chen, Q.; Shi, H.; Luo, X. Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction-induced myocardial damage via downregulation of early growth response factor 1. J. Cell. Biochem. 2018, 120, 4434–4443. [Google Scholar] [CrossRef] [PubMed]
- Raggi, P. Epicardial adipose tissue as a marker of coronary artery disease risk. J. Am. Coll. Cardiol. 2013, 61, 1396–1397. [Google Scholar] [CrossRef]
- De Gonzalo-Calvo, D.; Davalos, A.; Fernandez-Sanjurjo, M.; Amado-Rodriguez, L.; Diaz-Coto, S.; Tomas-Zapico, C.; Montero, A.; Garcia-Gonzalez, A.; Llorente-Cortes, V.; Heras, M.E.; et al. Circulating microRNAs as emerging cardiac biomarkers responsive to acute exercise. Int. J. Cardiol. 2018, 264, 130–136. [Google Scholar] [CrossRef]




| Variable | All | Tertile 1&2 | Tertile 3 | p Value |
|---|---|---|---|---|
| N = 180 | N = 120 | N = 60 | ||
| Clinical characteristics | ||||
| Age (years), mean ± SD | 65.0 ± 12.8 | 63.5 ± 13.8 | 68.1 ± 9.9 | 0.011 |
| Male, N (%) | 104 (58) | 63 (53) | 41 (68) | 0.055 |
| Body mass index (kg m−2), median (P25–P75) | 27.0 (24.8–30.3) | 25.9 (24.2–29.2) | 29.4 (26.3–32.1) | <0.001 |
| Body surface area (m2), median (P25–P75) N = 160 | 1.8 (1.7–2.0) | 1.8 (1.7–1.9) | 1.9 (1.9–2.1) | 0.001 |
| Hypertension, N (%) | 111 (62) | 70 (58) | 41 (68) | 0.255 |
| Dyslipidemia, N (%) | 102 (57) | 66 (55) | 36 (60) | 0.632 |
| Diabetes mellitus, N (%) | 37 (21) | 18 (15) | 19 (32) | 0.011 |
| Active or former smoker, N (%) | 59 (33) | 36 (30) | 23 (38) | 0.310 |
| hs-CRP (mg L−1), median (P25–P75) | 2.00 (0.97–4.07) | 1.90 (0.85–4.00) | 2.10 (1.11–4.62) | 0.590 |
| Coronary artery disease, N (%) | 55 (30.6) | 35 (29.2) | 20 (33.3) | 0.608 |
| Glomerular filtration rate < 60 mL/mi/1.73 m2, N (%) | 16 (9) | 11 (9) | 5 (8) | 1.000 |
| Medication use | ||||
| Antiplatelet drugs, N (%) | 73 (41) | 43 (36) | 40 (50) | 0.071 |
| Statins, N (%) | 87 (48) | 52 (43) | 35 (58) | 0.052 |
| Beta-blockers, N (%) | 58 (32) | 34 (28) | 24 (40) | 0.086 |
| Angiotensin-converting-enzyme inhibitors, N (%) | 97 (54) | 64 (53) | 33 (55) | 0.746 |
| Diuretics, N (%) | 48 (27) | 31 (26) | 17 (28) | 0.718 |
| Epicardial fat burden | ||||
| Epicardial fat volume (cm3), median (P25–P75) | 96.0 (66.5–130.6) | 79.3 (55.8–96.4) | 146.4 (130.5–178.4) | <0.001 |
| Epicardial fat volume-indexed (cm3 m−2), median (P25–P75) N = 160 | 50.0 (38.2–67.2) | 42.0 (32.1–52.4) | 76.3 (67.4–92.9) | <0.001 |
| Model 1 | Model 2 | Model 3 | Model 4 | |||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| miR-15b-3p | 1.701 (1.158–2.500) | 0.007 | 1.800 (1.177–2.753) | 0.007 | 1.832 (1.181–2.844) | 0.007 | 1.793 (1.174–2.738) | 0.007 |
| miR-15b-5p | 1.132 (0.907–1.412) | 0.273 | 1.183 (0..923–1.516) | 0.185 | 1.168 (0.905–1.507) | 0.234 | 1.182 (0.922–1.515) | 0.188 |
| miR-21-5p | 1.092 (0.824–1.446) | 0.540 | 1.169 (0.856–1.596) | 0.327 | 1.127 (0.816–1.555) | 0.468 | 1.168 (0.855–1.596) | 0.329 |
| miR-22-3p | 1.551 (1.075–2.239) | 0.019 | 1.677 (1.113–2.527) | 0.013 | 1.655 (1.089–2.516) | 0.018 | 1.669 (1.109–2.514) | 0.014 |
| miR-27a-3p | 1.100 (0.865–1.398) | 0.438 | 1.146 (0.880–1.492) | 0.311 | 1.120 (0.852–1.471) | 0.417 | 1.142 (0.877–1.488) | 0.324 |
| miR-27b-3p | 1.269 (0.976–1.651) | 0.076 | 1.331 (0.994–1.781) | 0.055 | 1.320 (0.977–1.782) | 0.070 | 1.325 (0.989–1.774) | 0.059 |
| miR-146a-5p | 1.010 (0.781–1.305) | 0.942 | 1.066 (0.805–1.410) | 0.657 | 1.027 (0.768–1.372) | 0.858 | 1.062 (0.801–1.406) | 0.677 |
| miR-148a–3p | 1.387 (1.052–1.829) | 0.020 | 1.417 (1.045–1.921) | 0.025 | 1.429 (1.045–1.955) | 0.025 | 1.417 (1.045–1.923) | 0.025 |
| miR-148b-3p | 1.444 (1.081–1.929) | 0.013 | 1.563 (1.130–2.161) | 0.007 | 1.527 (1.096–2.128) | 0.012 | 1.558 (1.127–2.154) | 0.007 |
| miR-152-3p | 1.222 (0.945–1.581) | 0.127 | 1.310 (0.982–1.748) | 0.066 | 1.283 (0.953–1.728) | 0.101 | 1.306 (0.979–1.744) | 0.070 |
| miR-339-3p | 1.304 (0.985–1.726) | 0.064 | 1.350 (0.992–1.838) | 0.056 | 1.317 (0.960–1.806) | 0.088 | 1.344 (0.988–1.830) | 0.060 |
| miR-590-5p | 1.449 (1.018–2.062) | 0.039 | 1.571 (1.062–2.324) | 0.024 | 1.541 (1.030–2.306) | 0.036 | 1.564 (1.059–2.312) | 0.025 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Gonzalo-Calvo, D.; Vilades, D.; Martínez-Camblor, P.; Vea, À.; Ferrero-Gregori, A.; Nasarre, L.; Bornachea, O.; Sanchez Vega, J.; Leta, R.; Puig, N.; et al. Plasma microRNA Profiling Reveals Novel Biomarkers of Epicardial Adipose Tissue: A Multidetector Computed Tomography Study. J. Clin. Med. 2019, 8, 780. https://doi.org/10.3390/jcm8060780
de Gonzalo-Calvo D, Vilades D, Martínez-Camblor P, Vea À, Ferrero-Gregori A, Nasarre L, Bornachea O, Sanchez Vega J, Leta R, Puig N, et al. Plasma microRNA Profiling Reveals Novel Biomarkers of Epicardial Adipose Tissue: A Multidetector Computed Tomography Study. Journal of Clinical Medicine. 2019; 8(6):780. https://doi.org/10.3390/jcm8060780
Chicago/Turabian Stylede Gonzalo-Calvo, David, David Vilades, Pablo Martínez-Camblor, Àngela Vea, Andreu Ferrero-Gregori, Laura Nasarre, Olga Bornachea, Jesus Sanchez Vega, Rubén Leta, Núria Puig, and et al. 2019. "Plasma microRNA Profiling Reveals Novel Biomarkers of Epicardial Adipose Tissue: A Multidetector Computed Tomography Study" Journal of Clinical Medicine 8, no. 6: 780. https://doi.org/10.3390/jcm8060780